7X4L
 
 | | Crystal structure of Bacteroides thetaiotaomicron glutamate decarboxylase mutant Y303F-PLP complex | | Descriptor: | Glutamate decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Liu, S, Guoming, D, Yulu, W, Boting, W, Xin, F. | | Deposit date: | 2022-03-02 | | Release date: | 2023-05-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Coordinated regulation of Bacteroides thetaiotaomicron glutamate decarboxylase activity by multiple elements under different pH.
Food Chem, 403, 2023
|
|
5UOO
 
 | | BRD4 bromodomain 2 in complex with CD161 | | Descriptor: | 7-(3,5-dimethyl-1,2-oxazol-4-yl)-6-methoxy-2-methyl-4-(quinolin-4-yl)-9H-pyrimido[4,5-b]indole, Bromodomain-containing protein 4 | | Authors: | Meagher, J.L, Stuckey, J.A. | | Deposit date: | 2017-02-01 | | Release date: | 2017-05-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structure-Based Discovery of 4-(6-Methoxy-2-methyl-4-(quinolin-4-yl)-9H-pyrimido[4,5-b]indol-7-yl)-3,5-dimethylisoxazole (CD161) as a Potent and Orally Bioavailable BET Bromodomain Inhibitor.
J. Med. Chem., 60, 2017
|
|
1C2W
 
 | |
3UF5
 
 | | Crystal structure of the mouse Colony-Stimulating Factor 1 (mCSF-1) cytokine | | Descriptor: | CALCIUM ION, Macrophage colony-stimulating factor 1 | | Authors: | Elegheert, J, Bracke, N, Bekaert, A, Savvides, S.N. | | Deposit date: | 2011-10-31 | | Release date: | 2012-08-22 | | Last modified: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric competitive inactivation of hematopoietic CSF-1 signaling by the viral decoy receptor BARF1
Nat.Struct.Mol.Biol., 19, 2012
|
|
3UEZ
 
 | |
2BE7
 
 | | Crystal structure of the unliganded (T-state) aspartate transcarbamoylase of the psychrophilic bacterium Moritella profunda | | Descriptor: | Aspartate Carbamoyltransferase Catalytic Chain, Aspartate Carbamoyltransferase Regulatory Chain, SULFATE ION, ... | | Authors: | De Vos, D, Savvides, S.N, Van Beeumen, J. | | Deposit date: | 2005-10-23 | | Release date: | 2006-10-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural investigation of cold activity and regulation of aspartate carbamoyltransferase from the extreme psychrophilic bacterium Moritella profunda.
J.Mol.Biol., 365, 2007
|
|
3QS9
 
 | |
3QS7
 
 | |
1E32
 
 | | Structure of the N-Terminal domain and the D1 AAA domain of membrane fusion ATPase p97 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, P97 | | Authors: | Zhang, X, Shaw, A, Bates, P.A, Gorman, M.A, Kondo, H, Dokurno, P, Leonard M, G, Sternberg, J.E, Freemont, P.S. | | Deposit date: | 2000-06-05 | | Release date: | 2001-05-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the Aaa ATPase P97
Mol.Cell, 6, 2000
|
|
7LV3
 
 | | Crystal structure of human protein kinase G (PKG) R-C complex in inhibited state | | Descriptor: | 1,2-ETHANEDIOL, Isoform Beta of cGMP-dependent protein kinase 1, MANGANESE (II) ION, ... | | Authors: | Sharma, R, Lying, Q, Casteel, D, Kim, C. | | Deposit date: | 2021-02-23 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | An auto-inhibited state of protein kinase G and implications for selective activation.
Elife, 11, 2022
|
|
7MBJ
 
 | |
4KU8
 
 | | Structures of PKGI Reveal a cGMP-Selective Activation Mechanism | | Descriptor: | GLYCINE, cGMP-dependent Protein Kinase 1 | | Authors: | Huang, G.Y, Kim, J.J, Reger, A.S, Lorenz, R, Moon, E.W, Casteel, D.E, Sankaran, B, Herberg, F.W, Kim, C. | | Deposit date: | 2013-05-21 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | Structural Basis for Cyclic-Nucleotide Selectivity and cGMP-Selective Activation of PKG I.
Structure, 22, 2014
|
|
4KU7
 
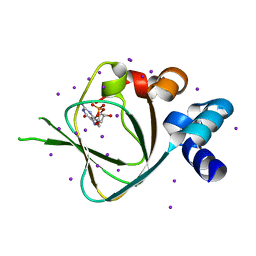 | | Structures of PKGI Reveal a cGMP-Selective Activation Mechanism | | Descriptor: | CYCLIC GUANOSINE MONOPHOSPHATE, IODIDE ION, cGMP-dependent protein kinase 1 | | Authors: | Huang, G.Y, Kim, J.J, Reger, A.S, Lorenz, R, Moon, E.W, Casteel, D.E, Sankaran, B, Herberg, F.W, Kim, C. | | Deposit date: | 2013-05-21 | | Release date: | 2014-01-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for Cyclic-Nucleotide Selectivity and cGMP-Selective Activation of PKG I.
Structure, 22, 2014
|
|
4ADF
 
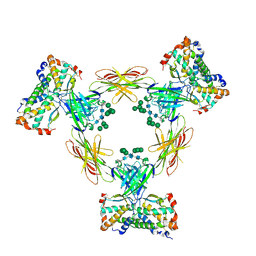 | | CRYSTAL STRUCTURE OF THE HUMAN COLONY-STIMULATING FACTOR 1 (hCSF-1) CYTOKINE IN COMPLEX WITH THE VIRAL RECEPTOR BARF1 | | Descriptor: | MACROPHAGE COLONY-STIMULATING FACTOR 1, SECRETED PROTEIN BARF1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Elegheert, J, Bracke, N, Savvides, S.N. | | Deposit date: | 2011-12-23 | | Release date: | 2012-08-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.4 Å) | | Cite: | Allosteric Competitive Inactivation of Hematopoietic Csf-1 Signaling by the Viral Decoy Receptor Barf1.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4ADQ
 
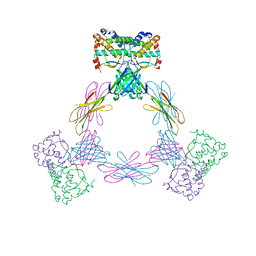 | | CRYSTAL STRUCTURE OF THE MOUSE COLONY-STIMULATING FACTOR 1 (MCSF-1) CYTOKINE IN COMPLEX WITH THE VIRAL RECEPTOR BARF1 | | Descriptor: | MACROPHAGE COLONY-STIMULATING FACTOR 1, SECRETED PROTEIN BARF1, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Elegheert, J, Bracke, N, Savvides, S.N. | | Deposit date: | 2012-01-02 | | Release date: | 2012-08-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Allosteric Competitive Inactivation of Hematopoietic Csf-1 Signaling by the Viral Decoy Receptor Barf1.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5D28
 
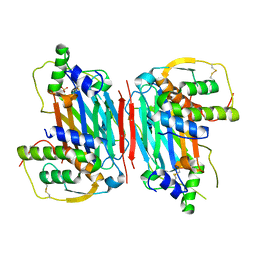 | |
5D22
 
 | |
3UF2
 
 | |
487D
 
 | |
8C7L
 
 | |
1C2X
 
 | |
1KWQ
 
 | | HUMAN CARBONIC ANHYDRASE II COMPLEXED WITH INHIBITOR 2000-07 | | Descriptor: | 3-NITRO-4-(2-OXO-PYRROLIDIN-1-YL)-BENZENESULFONAMIDE, Carbonic anhydrase II, MERCURY (II) ION, ... | | Authors: | Grueneberg, S, Stubbs, M.T. | | Deposit date: | 2002-01-30 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Successful virtual screening for novel inhibitors of human carbonic anhydrase: strategy and experimental confirmation.
J.Med.Chem., 45, 2002
|
|
1KWR
 
 | | HUMAN CARBONIC ANHYDRASE II COMPLEXED WITH INHIBITOR 0134-36 | | Descriptor: | 1-METHYL-3-OXO-1,3-DIHYDRO-BENZO[C]ISOTHIAZOLE-5-SULFONIC ACID AMIDE, Carbonic anhydrase II, ZINC ION | | Authors: | Grueneberg, S, Stubbs, M.T. | | Deposit date: | 2002-01-30 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Successful virtual screening for novel inhibitors of human carbonic anhydrase: strategy and experimental confirmation.
J.Med.Chem., 45, 2002
|
|
7K79
 
 | | CBF3 | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, Suppressor of kinetochore protein 1 | | Authors: | Ruifang, G, Yawen, B. | | Deposit date: | 2020-09-22 | | Release date: | 2021-03-31 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural and dynamic mechanisms of CBF3-guided centromeric nucleosome formation.
Nat Commun, 12, 2021
|
|
7K78
 
 | | antibody and nucleosome complex | | Descriptor: | Cse4, DNA (136-MER), Histone H2A.1, ... | | Authors: | Ruifang, G, Yawen, B. | | Deposit date: | 2020-09-22 | | Release date: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and dynamic mechanisms of CBF3-guided centromeric nucleosome formation.
Nat Commun, 12, 2021
|
|
