4Z6S
 
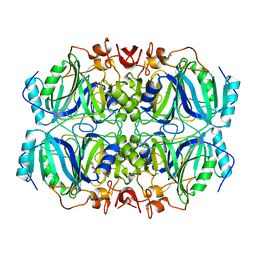 | | Structure of H200Q variant of Homoprotocatechuate 2,3-Dioxygenase from B.fuscum in complex with 4-sulfonyl catechol at 1.42 Ang resolution | | Descriptor: | 3,4-dihydroxybenzenesulfonic acid, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kovaleva, E.G, Lipscomb, J.D. | | Deposit date: | 2015-04-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Structural Basis for Substrate and Oxygen Activation in Homoprotocatechuate 2,3-Dioxygenase: Roles of Conserved Active Site Histidine 200.
Biochemistry, 54, 2015
|
|
4Z6U
 
 | | Structure of H200E variant of Homoprotocatechuate 2,3-Dioxygenase from B.fuscum in complex with 4-nitrocatechol at 1.48 Ang resolution | | Descriptor: | 4-NITROCATECHOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kovaleva, E.G, Lipscomb, J.D. | | Deposit date: | 2015-04-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structural Basis for Substrate and Oxygen Activation in Homoprotocatechuate 2,3-Dioxygenase: Roles of Conserved Active Site Histidine 200.
Biochemistry, 54, 2015
|
|
4Z6W
 
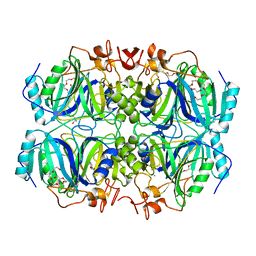 | | Structure of H200N variant of Homoprotocatechuate 2,3-Dioxygenase from B.fuscum in complex with 4-nitrocatechol at 1.57 Ang resolution | | Descriptor: | 4-NITROCATECHOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kovaleva, E.G, Lipscomb, J.D. | | Deposit date: | 2015-04-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural Basis for Substrate and Oxygen Activation in Homoprotocatechuate 2,3-Dioxygenase: Roles of Conserved Active Site Histidine 200.
Biochemistry, 54, 2015
|
|
4Z6P
 
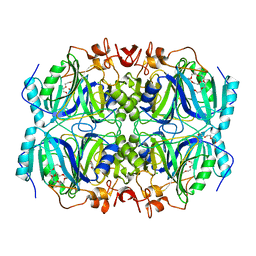 | | Structure of H200Q variant of Homoprotocatechuate 2,3-Dioxygenase from B.fuscum in complex with HPCA at 1.75 Ang resolution | | Descriptor: | 2-(3,4-DIHYDROXYPHENYL)ACETIC ACID, CHLORIDE ION, FE (II) ION, ... | | Authors: | Kovaleva, E.G, Lipscomb, J.D. | | Deposit date: | 2015-04-06 | | Release date: | 2015-08-26 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Substrate and Oxygen Activation in Homoprotocatechuate 2,3-Dioxygenase: Roles of Conserved Active Site Histidine 200.
Biochemistry, 54, 2015
|
|
5BWG
 
 | | Structure of H200C variant of Homoprotocatechuate 2,3-Dioxygenase from B.fuscum at 1.75 Ang resolution | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kovaleva, E.G, Lipscomb, J.D. | | Deposit date: | 2015-06-08 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A Long-Lived Fe(III)-(Hydroperoxo) Intermediate in the Active H200C Variant of Homoprotocatechuate 2,3-Dioxygenase: Characterization by Mossbauer, Electron Paramagnetic Resonance, and Density Functional Theory Methods.
Inorg.Chem., 54, 2015
|
|
5BWH
 
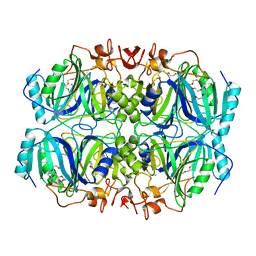 | | Structure of H200C variant of Homoprotocatechuate 2,3-Dioxygenase from B.fuscum in complex with HPCA at 1.46 Ang resolution | | Descriptor: | 2-(3,4-DIHYDROXYPHENYL)ACETIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Kovaleva, E.G, Lipscomb, J.D. | | Deposit date: | 2015-06-08 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | A Long-Lived Fe(III)-(Hydroperoxo) Intermediate in the Active H200C Variant of Homoprotocatechuate 2,3-Dioxygenase: Characterization by Mossbauer, Electron Paramagnetic Resonance, and Density Functional Theory Methods.
Inorg.Chem., 54, 2015
|
|
1Q0C
 
 | | Anerobic Substrate Complex of Homoprotocatechuate 2,3-Dioxygenase from Brevibacterium fuscum. (Complex with 3,4-Dihydroxyphenylacetate) | | Descriptor: | 2-(3,4-DIHYDROXYPHENYL)ACETIC ACID, FE (III) ION, homoprotocatechuate 2,3-dioxygenase | | Authors: | Vetting, M.W, Wackett, L.P, Que, L, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2003-07-15 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic comparison of manganese- and iron-dependent homoprotocatechuate 2,3-dioxygenases.
J.Bacteriol., 186, 2004
|
|
1Q0O
 
 | | CRYSTAL STRUCTURE OF HOMOPROTOCATECHUATE 2,3-DIOXYGENASE FROM BREVIBACTERIUM FUSCUM (FULL LENGTH PROTEIN) | | Descriptor: | FE (III) ION, homoprotocatechuate 2,3-dioxygenase | | Authors: | Vetting, M.W, Wackett, L.P, Que, L, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 2003-07-16 | | Release date: | 2003-07-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic comparison of manganese- and iron-dependent homoprotocatechuate 2,3-dioxygenases.
J.Bacteriol., 186, 2004
|
|
1F1R
 
 | | CRYSTAL STRUCTURE OF HOMOPROTOCATECHUATE 2,3-DIOXYGENASE FROM ARTHROBACTER GLOBIFORMIS (NATIVE, NON-CRYO) | | Descriptor: | HOMOPROTOCATECHUATE 2,3-DIOXYGENASE, MANGANESE (II) ION | | Authors: | Vetting, M.W, Lipscomb, J.D, Wackett, L.P, Que Jr, L, Ohlendorf, D.H. | | Deposit date: | 2000-05-19 | | Release date: | 2003-06-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic comparison of manganese- and iron-dependent homoprotocatechuate 2,3-dioxygenases.
J.Bacteriol., 186, 2004
|
|
1F1V
 
 | | ANAEROBIC SUBSTRATE COMPLEX OF HOMOPROTOCATECHUATE 2,3-DIOXYGENASE FROM ARTHROBACTER GLOBIFORMIS. (COMPLEX WITH 3,4-DIHYDROXYPHENYLACETATE) | | Descriptor: | 2-(3,4-DIHYDROXYPHENYL)ACETIC ACID, HOMOPROTOCATECHUATE 2,3-DIOXYGENASE, MANGANESE (II) ION | | Authors: | Vetting, M.W, Lipscomb, J.D, Wackett, L.P, Que Jr, L, Ohlendorf, D.H. | | Deposit date: | 2000-05-20 | | Release date: | 2003-06-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic comparison of manganese- and iron-dependent homoprotocatechuate 2,3-dioxygenases.
J.Bacteriol., 186, 2004
|
|
1F1U
 
 | | CRYSTAL STRUCTURE OF HOMOPROTOCATECHUATE 2,3-DIOXYGENASE FROM ARTHROBACTER GLOBIFORMIS (NATIVE, LOW TEMPERATURE) | | Descriptor: | HOMOPROTOCATECHUATE 2,3-DIOXYGENASE, MANGANESE (II) ION | | Authors: | Vetting, M.W, Lipscomb, J.D, Wackett, L.P, Que Jr, L, Ohlendorf, D.H. | | Deposit date: | 2000-05-19 | | Release date: | 2003-06-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystallographic comparison of manganese- and iron-dependent homoprotocatechuate 2,3-dioxygenases.
J.Bacteriol., 186, 2004
|
|
7Q2U
 
 | | The crystal structure of the HINT1 Q62A mutant. | | Descriptor: | CACODYLATE ION, HEXAETHYLENE GLYCOL, Histidine triad nucleotide-binding protein 1, ... | | Authors: | Dolot, R.M, Strom, A.M, Wagner, C.R. | | Deposit date: | 2021-10-26 | | Release date: | 2021-11-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Dynamic Long-Range Interactions Influence Substrate Binding and Catalysis by Human Histidine Triad Nucleotide-Binding Proteins (HINTs), Key Regulators of Multiple Cellular Processes and Activators of Antiviral ProTides.
Biochemistry, 61, 2022
|
|
5MND
 
 | |
6YD0
 
 | | XFEL structure of the Soluble methane monooxygenase hydroxylase and regulatory subunit complex, from Methylosinus trichosporium OB3b, diferric state | | Descriptor: | FE (III) ION, GLYCEROL, Methane monooxygenase, ... | | Authors: | Srinivas, V, Hogbom, M. | | Deposit date: | 2020-03-19 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-Resolution XFEL Structure of the Soluble Methane Monooxygenase Hydroxylase Complex with its Regulatory Component at Ambient Temperature in Two Oxidation States.
J. Am. Chem. Soc., 142, 2020
|
|
6YDU
 
 | | XFEL structure of the Soluble methane monooxygenase hydroxylase and regulatory subunit complex, from Methylosinus trichosporium OB3b, reoxidized diferric state, 10s O2 exposure. | | Descriptor: | FE (III) ION, GLYCEROL, Methane monooxygenase, ... | | Authors: | Srinivas, V, Hogbom, M. | | Deposit date: | 2020-03-21 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-Resolution XFEL Structure of the Soluble Methane Monooxygenase Hydroxylase Complex with its Regulatory Component at Ambient Temperature in Two Oxidation States.
J. Am. Chem. Soc., 142, 2020
|
|
5G5J
 
 | | Crystal structure of human CYP3A4 bound to metformin | | Descriptor: | CYTOCHROME P450 3A4, Metformin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I. | | Deposit date: | 2016-05-25 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Heme Binding Biguanides Target Cytochrome P450-Dependent Cancer Cell Mitochondria.
Cell Chem Biol, 24, 2017
|
|
6YDI
 
 | | XFEL structure of the Soluble methane monooxygenase hydroxylase and regulatory subunit complex, from Methylosinus trichosporium OB3b, diferrous state | | Descriptor: | FE (II) ION, GLYCEROL, Methane monooxygenase, ... | | Authors: | Srinivas, V, Hogbom, M. | | Deposit date: | 2020-03-20 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-Resolution XFEL Structure of the Soluble Methane Monooxygenase Hydroxylase Complex with its Regulatory Component at Ambient Temperature in Two Oxidation States.
J. Am. Chem. Soc., 142, 2020
|
|
6YY3
 
 | | XFEL structure of the Soluble methane monooxygenase hydroxylase and regulatory subunit complex, from Methylosinus trichosporium OB3b, t=0 diferrous state prior to oxygen activation | | Descriptor: | FE (II) ION, GLYCEROL, Methane monooxygenase, ... | | Authors: | Srinivas, V, Hogbom, M. | | Deposit date: | 2020-05-04 | | Release date: | 2020-09-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-Resolution XFEL Structure of the Soluble Methane Monooxygenase Hydroxylase Complex with its Regulatory Component at Ambient Temperature in Two Oxidation States.
J. Am. Chem. Soc., 142, 2020
|
|
5TE2
 
 | | Crystal structure of 7,8-diaminopelargonic acid synthase (BioA) from Mycobacterium tuberculosis, complexed with a mechanism-based inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Maize, K.M, Eiden, C, Aldrich, C.C, Finzel, B.C. | | Deposit date: | 2016-09-20 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rational Optimization of Mechanism-Based Inhibitors through Determination of the Microscopic Rate Constants of Inactivation.
J. Am. Chem. Soc., 139, 2017
|
|
5U5Q
 
 | | 12 Subunit RNA Polymerase II at Room Temperature collected using SFX | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Bushnell, D.A, Oberthur, D, Mariani, V, Yefanov, O, Tolstikova, A, Barty, A. | | Deposit date: | 2016-12-07 | | Release date: | 2017-03-29 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Double-flow focused liquid injector for efficient serial femtosecond crystallography.
Sci Rep, 7, 2017
|
|
5TRX
 
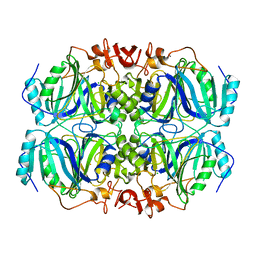 | | Room temperature structure of an extradiol ring-cleaving dioxygenase from B.fuscum determined using serial femtosecond crystallography | | Descriptor: | CALCIUM ION, CHLORIDE ION, FE (II) ION, ... | | Authors: | Kovaleva, E.G, Oberthuer, D, Tolstikova, A, Mariani, V. | | Deposit date: | 2016-10-27 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Double-flow focused liquid injector for efficient serial femtosecond crystallography
Sci. Rep., 7, 2017
|
|
3V5Z
 
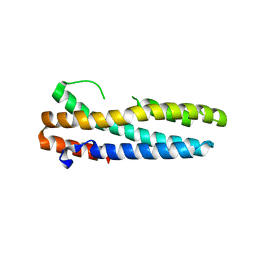 | | Structure of FBXL5 hemerythrin domain, C2 cell, grown anaerobically | | Descriptor: | F-box/LRR-repeat protein 5, MU-OXO-DIIRON | | Authors: | Tomchick, D.R, Bruick, R.K, Thompson, J.W, Brautigam, C.A. | | Deposit date: | 2011-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1847 Å) | | Cite: | Structural and Molecular Characterization of Iron-sensing Hemerythrin-like Domain within F-box and Leucine-rich Repeat Protein 5 (FBXL5).
J.Biol.Chem., 287, 2012
|
|
3V5Y
 
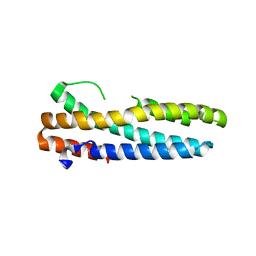 | | Structure of FBXL5 hemerythrin domain, P2(1) cell | | Descriptor: | F-box/LRR-repeat protein 5, MU-OXO-DIIRON | | Authors: | Tomchick, D.R, Bruick, R.K, Thompson, J.W, Brautigam, C.A. | | Deposit date: | 2011-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Molecular Characterization of Iron-sensing Hemerythrin-like Domain within F-box and Leucine-rich Repeat Protein 5 (FBXL5).
J.Biol.Chem., 287, 2012
|
|
3V5X
 
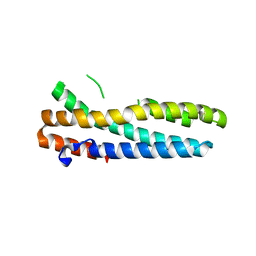 | | Structure of FBXL5 hemerythrin domain, C2 cell | | Descriptor: | F-box/LRR-repeat protein 5, MU-OXO-DIIRON | | Authors: | Tomchick, D.R, Bruick, R.K, Thompson, J.W, Brautigam, C.A. | | Deposit date: | 2011-12-17 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural and Molecular Characterization of Iron-sensing Hemerythrin-like Domain within F-box and Leucine-rich Repeat Protein 5 (FBXL5).
J.Biol.Chem., 287, 2012
|
|
4E37
 
 | | Crystal Structure of P. aeruginosa catalase, KatA tetramer | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | VanderWielen, B.D, Wilson, J.J, Kovall, R.A. | | Deposit date: | 2012-03-09 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | KatA, the Major Catalase in Pseudomonas aeruginosa, Assists in Protecting Anaerobic Bacteria From Metabolic and Exogenous Nitric Oxide
To be Published
|
|
