1GSF
 
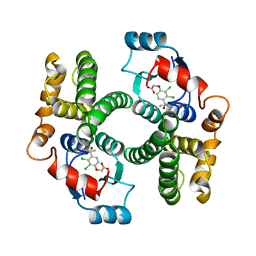 | | GLUTATHIONE TRANSFERASE A1-1 COMPLEXED WITH ETHACRYNIC ACID | | Descriptor: | ETHACRYNIC ACID, GLUTATHIONE TRANSFERASE A1-1 | | Authors: | L'Hermite, G, Sinning, I, Cameron, A.D, Jones, T.A. | | Deposit date: | 1995-06-09 | | Release date: | 1995-09-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural analysis of human alpha-class glutathione transferase A1-1 in the apo-form and in complexes with ethacrynic acid and its glutathione conjugate.
Structure, 3, 1995
|
|
1GSD
 
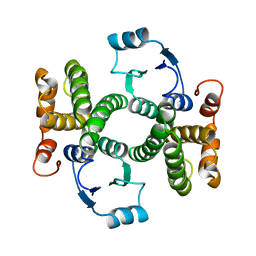 | |
3ZPP
 
 | |
4USC
 
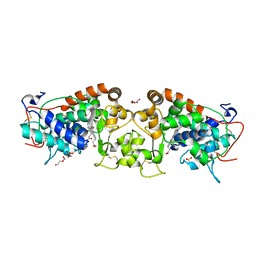 | | Crystal structure of peroxidase from palm tree Chamaerops excelsa | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Bernardes, A, Santos, J.C, Textor, L.C, Cuadrado, N.H, Kostetsky, E.Y, Roig, M.G, Muniz, J.R.C, Shnyrov, V.L, Polikarpov, I. | | Deposit date: | 2014-07-07 | | Release date: | 2015-05-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure Analysis of Peroxidase from the Palm Tree Chamaerops Excelsa.
Biochimie, 111, 2015
|
|
4UW7
 
 | | Structure of the carboxy-terminal domain of the bacteriophage T5 L- shaped tail fiber without its intra-molecular chaperone domain | | Descriptor: | GLYCEROL, L-SHAPED TAIL FIBER PROTEIN | | Authors: | Garcia-Doval, C, Luque, D, Caston, J.R, Otero, J.M, Llamas-Saiz, A.L, Boulanger, P, van Raaij, M.J. | | Deposit date: | 2014-08-08 | | Release date: | 2015-08-05 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structure of the Receptor-Binding Carboxy-Terminal Domain of the Bacteriophage T5 L-Shaped Tail Fibre with and without Its Intra-Molecular Chaperone.
Viruses, 7, 2015
|
|
4UY8
 
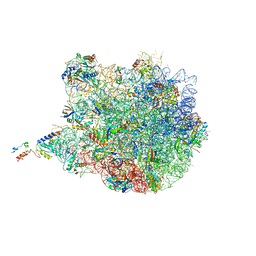 | | Molecular basis for the ribosome functioning as a L-tryptophan sensor - Cryo-EM structure of a TnaC stalled E.coli ribosome | | Descriptor: | 50S RIBOSOMAL PROTEIN L10, 50S RIBOSOMAL PROTEIN L11, 50S RIBOSOMAL PROTEIN L13, ... | | Authors: | Bischoff, L, Berninghausen, O, Beckmann, R. | | Deposit date: | 2014-08-29 | | Release date: | 2014-10-29 | | Last modified: | 2018-10-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular Basis for the Ribosome Functioning as an L-Tryptophan Sensor.
Cell Rep., 9, 2014
|
|
4UQH
 
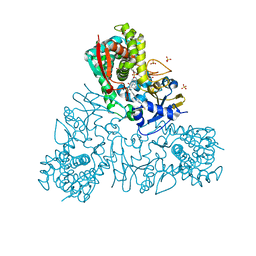 | | Crystal structure of Trypanosoma cruzi CYP51 bound to the inhibitor (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-4-(4-(3,4-difluorophenyl)piperazin-1-yl)-2-fluorobenzamide. | | Descriptor: | (R)-N-(3-(1H-indol-3-yl)-1-oxo-1-(pyridin-4-ylamino)propan-2-yl)-4-(4-(3,4-difluorophenyl)piperazin-1-yl)-2-fluorobenzamide, PROTOPORPHYRIN IX CONTAINING FE, STEROL 14-ALPHA DEMETHYLASE, ... | | Authors: | Calvet, C.M, Vieira, D.F, Choi, J.Y, Cameron, M.D, Gut, J, Kellar, D, Siqueira-Neto, J.L, McKerrow, J.H, Roush, W.R, Podust, L.M. | | Deposit date: | 2014-06-23 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | 4-Aminopyridyl-Based Cyp51 Inhibitors as Anti-Trypanosoma Cruzi Drug Leads with Improved Pharmacokinetic Profile and in Vivo Potency.
J.Med.Chem., 57, 2014
|
|
4V3Y
 
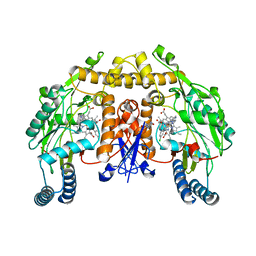 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with N-2-(2-(1H-imidazol-1-yl)pyrimidin-4-yl)ethyl-3-(3- chlorophenyl)propan-1-amine | | Descriptor: | 3-(3-chlorophenyl)-N-{2-[2-(1H-imidazol-1-yl)pyrimidin-4-yl]ethyl}propan-1-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2014-10-20 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.962 Å) | | Cite: | Novel 2,4-Disubstituted Pyrimidines as Potent, Selective, and Cell-Permeable Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
4V3V
 
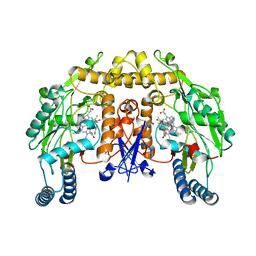 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with N-(2-(1H-imidazol-1-yl)-4-pyrimidylmethyl)-3-(3- fluorophenyl)propan-1-amine | | Descriptor: | 3-(3-fluorophenyl)-N-{[2-(1H-imidazol-1-yl)pyrimidin-4-yl]methyl}propan-1-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2014-10-20 | | Release date: | 2014-12-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Novel 2,4-Disubstituted Pyrimidines as Potent, Selective, and Cell-Permeable Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
4V3U
 
 | | Structure of human nNOS R354A G357D mutant heme domain in complex with N-2-(2-(1H-imidazol-1-yl)pyrimidin-4-yl)ethyl-3-(pyridin-3-yl) propan-1-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, GLYCEROL, N-{2-[2-(1H-imidazol-1-yl)pyrimidin-4-yl]ethyl}-3-(pyridin-3-yl)propan-1-amine, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2014-10-20 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel 2,4-Disubstituted Pyrimidines as Potent, Selective, and Cell-Permeable Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
4V9R
 
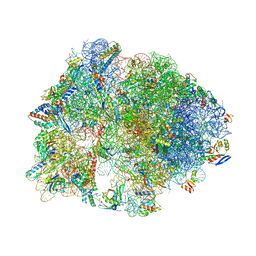 | | Crystal structure of antibiotic DITYROMYCIN bound to 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Bulkley, D.P, Brandi, L, Polikanov, Y.S, Fabbretti, A, O'Connor, M, Gualerzi, C.O, Steitz, T.A. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation.
Cell Rep, 6, 2014
|
|
7O84
 
 | | Structure of the PL6 family alginate lyase Pedsa0632 from Pseudopedobacter saltans in complex with substrate | | Descriptor: | 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid, 4-deoxy-alpha-L-erythro-hex-4-enopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid, Alginate lyase | | Authors: | Ballut, L, Violot, S, Carrique, L, Aghajari, N. | | Deposit date: | 2021-04-14 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.177 Å) | | Cite: | Exploring molecular determinants of polysaccharide lyase family 6-1 enzyme activity.
Glycobiology, 31, 2021
|
|
4UWQ
 
 | | Crystal structure of the disulfide-linked complex of the thiosulfodyrolase SoxB with the carrier-protein SoxYZ from Thermus thermophilus | | Descriptor: | MANGANESE (II) ION, SOXY PROTEIN, SOXZ, ... | | Authors: | Grabarczyk, D.B, Chappell, P.E, Johnson, S, Stelzl, L.S, Lea, S.M, Berks, B.C. | | Deposit date: | 2014-08-14 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.28 Å) | | Cite: | Structural Basis for Specificity and Promiscuity in a Carrier Protein/Enzyme System from the Sulfur Cycle
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4W2H
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with pactamycin (co-crystallized), mRNA and deacylated tRNA in the P site | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Polikanov, Y.S, Osterman, I.A, Szal, T, Tashlitsky, V.N, Serebryakova, M.V, Kusochek, P, Bulkley, D, Malanicheva, I.A, Efimenko, T.A, Efremenkova, O.V, Konevega, A.L, Shaw, K.J, Bogdanov, A.A, Rodnina, M.V, Dontsova, O.A, Mankin, A.S, Steitz, T.A, Sergiev, P.V. | | Deposit date: | 2014-09-12 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Amicoumacin a inhibits translation by stabilizing mRNA interaction with the ribosome.
Mol.Cell, 56, 2014
|
|
4UVW
 
 | | Crystal structure of human tankyrase 2 in complex with 4,5-dimethyl-3- phenyl-1,2-dihydroisoquinolin-1-one | | Descriptor: | 4,5-dimethyl-3-phenylisoquinolin-1(2H)-one, SULFATE ION, TANKYRASE-2, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2014-08-08 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
4WDY
 
 | | JC Polyomavirus VP1 five-fold pore mutant N221Q | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Major capsid protein VP1 | | Authors: | Stroh, L.J, Stehle, T. | | Deposit date: | 2014-09-09 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of a Pore in the Capsid of JC Polyomavirus Reduces Infectivity and Prevents Exposure of the Minor Capsid Proteins.
J.Virol., 89, 2015
|
|
4UT1
 
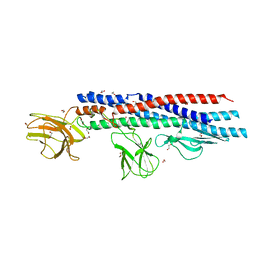 | |
4V25
 
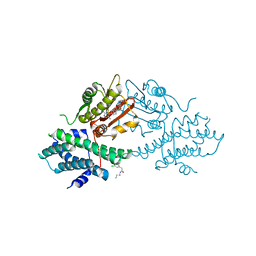 | | VER-246608, a novel pan-isoform ATP competitive inhibitor of pyruvate dehydrogenase kinase, disrupts Warburg metabolism and induces context- dependent cytostasis in cancer cells | | Descriptor: | MAGNESIUM ION, N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, N-[4-(2-chloro-5-methylpyrimidin-4-yl)phenyl]-N-(4-{[(difluoroacetyl)amino]methyl}benzyl)-2,4-dihydroxybenzamide, ... | | Authors: | Moore, J.D, Staniszewska, A, Shaw, T, D'Alessandro, J, Davis, B, Surgenor, A, Baker, L, Matassova, N, Murray, J, Macias, A, Brough, P, Wood, M, Mahon, P.C. | | Deposit date: | 2014-10-06 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | VER-246608, a novel pan-isoform ATP competitive inhibitor of pyruvate dehydrogenase kinase, disrupts Warburg metabolism and induces context-dependent cytostasis in cancer cells.
Oncotarget, 5, 2014
|
|
4V4P
 
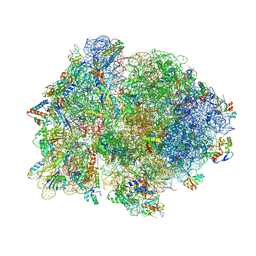 | | Crystal structure of 70S ribosome with thrS operator and tRNAs. | | Descriptor: | 16S rRNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Jenner, L, Romby, P, Rees, B, Schulze-Briese, C, Springer, M, Ehresmann, C, Ehresmann, B, Moras, D, Yusupova, G, Yusupov, M. | | Deposit date: | 2005-01-19 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Translational operator of mRNA on the ribosome: how repressor proteins exclude ribosome binding.
Science, 308, 2005
|
|
4V76
 
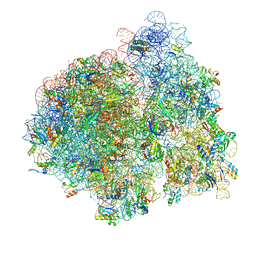 | | E. coli 70S-fMetVal-tRNAVal-tRNAfMet complex in intermediate post-translocation state (post2a) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Blau, C, Bock, L.V, Schroder, G.F, Davydov, I, Fischer, N, Stark, H, Rodnina, M.V, Vaiana, A.C, Grubmuller, H. | | Deposit date: | 2013-10-14 | | Release date: | 2014-07-09 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Energy barriers and driving forces in tRNA translocation through the ribosome.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4V87
 
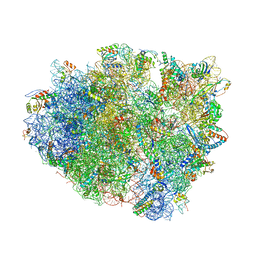 | | Crystal structure analysis of ribosomal decoding. | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Demeshkina, N, Jenner, L, Yusupov, M, Yusupova, G. | | Deposit date: | 2011-09-20 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
4UVU
 
 | | Crystal structure of human tankyrase 2 in complex with 1-((4-(5- methyl-1-oxo-1,2-dihydroisoquinolin-3-yl)phenyl)methyl)pyrrolidin-1- ium | | Descriptor: | 5-methyl-3-[4-(pyrrolidin-1-ylmethyl)phenyl]isoquinolin-1(2H)-one, GLYCEROL, SULFATE ION, ... | | Authors: | Haikarainen, T, Narwal, M, Lehtio, L. | | Deposit date: | 2014-08-08 | | Release date: | 2015-07-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
4UVS
 
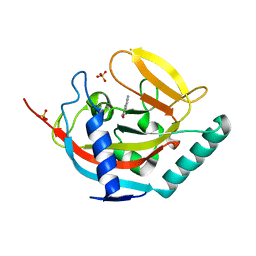 | | Crystal structure of human tankyrase 2 in complex with 5-amino-3- pentyl-1,2-dihydroisoquinolin-1-one | | Descriptor: | 5-amino-3-pentylisoquinolin-1(2H)-one, SULFATE ION, TANKYRASE-2, ... | | Authors: | Narwal, M, Haikarainen, T, Lehtio, L. | | Deposit date: | 2014-08-08 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Exploration of the Nicotinamide-Binding Site of the Tankyrases, Identifying 3-Arylisoquinolin-1-Ones as Potent and Selective Inhibitors in Vitro.
Bioorg.Med.Chem., 23, 2015
|
|
7O19
 
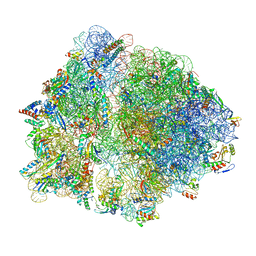 | | Cryo-EM structure of an Escherichia coli TnaC-ribosome complex stalled in response to L-tryptophan | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | van der Stel, A.X, Gordon, E.R, Sengupta, A, Martinez, A.K, Klepacki, D, Perry, T.N, Herrero del Valle, A, Vazquez-Laslop, N, Sachs, M.S, Cruz-Vera, L.R, Innis, C.A. | | Deposit date: | 2021-03-29 | | Release date: | 2021-09-01 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis for the tryptophan sensitivity of TnaC-mediated ribosome stalling.
Nat Commun, 12, 2021
|
|
7O1A
 
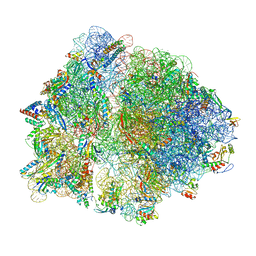 | | Cryo-EM structure of an Escherichia coli TnaC(R23F)-ribosome complex stalled in response to L-tryptophan | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | van der Stel, A.X, Gordon, E.R, Sengupta, A, Martinez, A.K, Klepacki, D, Perry, T.N, Herrero del Valle, A, Vazquez-Laslop, N, Sachs, M.S, Cruz-Vera, L.R, Innis, C.A. | | Deposit date: | 2021-03-29 | | Release date: | 2021-09-01 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural basis for the tryptophan sensitivity of TnaC-mediated ribosome stalling.
Nat Commun, 12, 2021
|
|
