3EIH
 
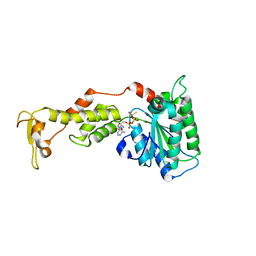 | | Crystal structure of S.cerevisiae Vps4 in the presence of ATPgammaS | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, ... | | Authors: | Gonciarz, M.D, Whitby, F.G, Eckert, D.M, Kieffer, C, Heroux, A, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2008-09-15 | | Release date: | 2008-09-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Biochemical and structural studies of yeast vps4 oligomerization.
J.Mol.Biol., 384, 2008
|
|
3LL5
 
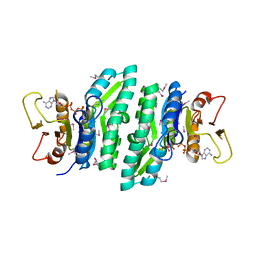 | |
3FRS
 
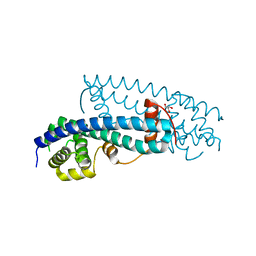 | | Structure of human IST1(NTD) (residues 1-189)(p43212) | | Descriptor: | GLYCEROL, Uncharacterized protein KIAA0174 | | Authors: | Schubert, H.L, Hill, C.P, Bajorek, M, Sundquist, W.I. | | Deposit date: | 2009-01-08 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis for ESCRT-III protein autoinhibition.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3L35
 
 | | PIE12 D-peptide against HIV entry | | Descriptor: | GP41 N-PEPTIDE, HIV ENTRY INHIBITOR PIE12 | | Authors: | Welch, B.D, Redman, J.S, Paul, S, Whitby, F.G, Weinstock, M.T, Reeves, J.D, Lie, Y.S, Eckert, D.M, Hill, C.P, Root, M.J, Kay, M.S. | | Deposit date: | 2009-12-16 | | Release date: | 2010-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design of a potent D-peptide HIV-1 entry inhibitor with a strong barrier to resistance.
J.Virol., 84, 2010
|
|
3LKK
 
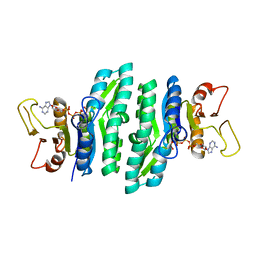 | |
6AP1
 
 | | Vps4p-Vta1p complex with peptide binding to the central pore of Vps4p | | Descriptor: | ACE-ASP-GLU-ILE-VAL-ASN-LYS-VAL-LEU-NH2, ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Han, H, Monroe, N, Shen, P, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2017-08-16 | | Release date: | 2017-12-06 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The AAA ATPase Vps4 binds ESCRT-III substrates through a repeating array of dipeptide-binding pockets.
Elife, 6, 2017
|
|
3O8Z
 
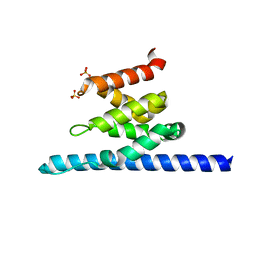 | | Crystal structure of Spn1 (Iws1) core domain | | Descriptor: | SULFATE ION, Transcription factor IWS1 | | Authors: | McDonald, S.M, Close, D, Hill, C.P. | | Deposit date: | 2010-08-03 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and biological importance of the spn1-spt6 interaction, and its regulatory role in nucleosome binding.
Mol.Cell, 40, 2010
|
|
2IDX
 
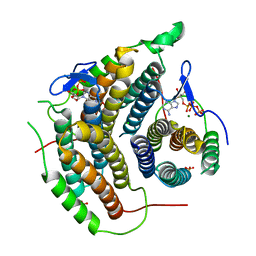 | |
3OAK
 
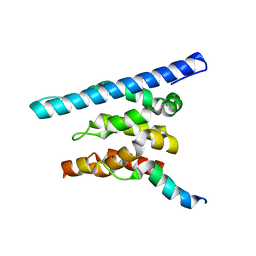 | | Crystal structure of a Spn1 (Iws1)-Spt6 complex | | Descriptor: | Transcription elongation factor SPT6, Transcription factor IWS1 | | Authors: | McDonald, S.M, Close, D, Hill, C.P. | | Deposit date: | 2010-08-05 | | Release date: | 2010-11-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure and biological importance of the spn1-spt6 interaction, and its regulatory role in nucleosome binding.
Mol.Cell, 40, 2010
|
|
3NWH
 
 | | Crystal structure of BST2/Tetherin | | Descriptor: | Bone marrow stromal antigen 2 | | Authors: | Schubert, H.L, Zhai, Q, Hill, C.P. | | Deposit date: | 2010-07-09 | | Release date: | 2010-07-21 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional studies on the extracellular domain of BST2/tetherin in reduced and oxidized conformations.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
6BMF
 
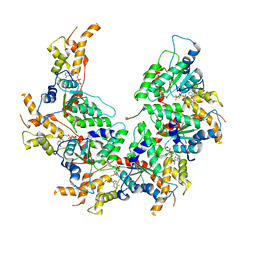 | | Vps4p-Vta1p complex with peptide binding to the central pore of Vps4p | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Han, H, Monroe, N, Shen, P, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2017-11-14 | | Release date: | 2017-12-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The AAA ATPase Vps4 binds ESCRT-III substrates through a repeating array of dipeptide-binding pockets.
Elife, 6, 2017
|
|
5V1Y
 
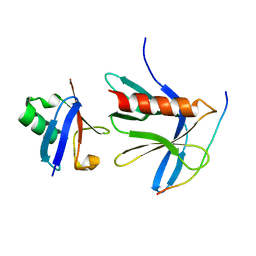 | | Crystal structure of the ternary RPN13 PRU-RPN2 (940-953)-ubiquitin complex | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Hemmis, C.W, VanderLinden, R.T, Yao, T, Robinson, H, Hill, C.P. | | Deposit date: | 2017-03-02 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.421 Å) | | Cite: | Structure and energetics of pairwise interactions between proteasome subunits RPN2, RPN13, and ubiquitin clarify a substrate recruitment mechanism.
J. Biol. Chem., 292, 2017
|
|
2HTH
 
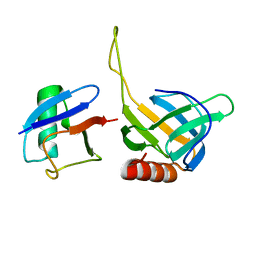 | | Structural basis for ubiquitin recognition by the human EAP45/ESCRT-II GLUE domain | | Descriptor: | Ubiquitin, Vacuolar protein sorting protein 36 | | Authors: | Alam, S.L, Whitby, F.G, Hill, C.P, Sundquist, W.I. | | Deposit date: | 2006-07-25 | | Release date: | 2006-10-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for ubiquitin recognition by the human ESCRT-II EAP45 GLUE domain.
Nat.Struct.Mol.Biol., 13, 2006
|
|
5UIE
 
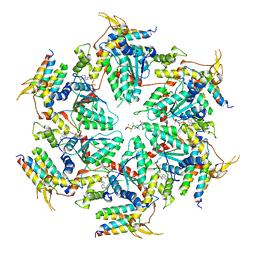 | | Vps4-Vta1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DOA4-independent degradation protein 4, ... | | Authors: | Monroe, N, Shen, P, Han, H, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2017-01-13 | | Release date: | 2017-04-12 | | Last modified: | 2020-01-01 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Structural basis of protein translocation by the Vps4-Vta1 AAA ATPase.
Elife, 6, 2017
|
|
2Q6Z
 
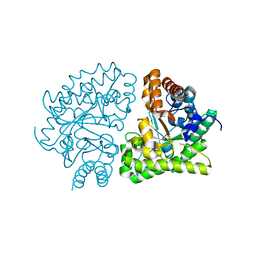 | | Uroporphyrinogen Decarboxylase G168R single mutant apo-enzyme | | Descriptor: | Uroporphyrinogen decarboxylase | | Authors: | Phillips, J.D, Whitby, F.G, Stadtmueller, B.M, Edwards, C.Q, Hill, C.P, Kushner, J.P. | | Deposit date: | 2007-06-05 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two novel uroporphyrinogen decarboxylase (URO-D) mutations causing hepatoerythropoietic porphyria (HEP).
Transl.Res., 149, 2007
|
|
5V1Z
 
 | | Crystal structure of the RPN13 PRU-RPN2 (932-953)-ubiquitin complex | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Hemmis, C.W, VanderLinden, R.T, Yao, T, Robinson, H, Hill, C.P. | | Deposit date: | 2017-03-02 | | Release date: | 2017-05-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and energetics of pairwise interactions between proteasome subunits RPN2, RPN13, and ubiquitin clarify a substrate recruitment mechanism.
J. Biol. Chem., 292, 2017
|
|
3PSJ
 
 | |
3PSI
 
 | |
2JPR
 
 | | Joint refinement of the HIV-1 CA-NTD in complex with the assembly inhibitor CAP-1 | | Descriptor: | 1-(3-chloro-4-methylphenyl)-3-{2-[({5-[(dimethylamino)methyl]-2-furyl}methyl)thio]ethyl}urea, Gag-Pol polyprotein | | Authors: | Kelly, B.N, Kyere, S, Kinde, I, Tang, C, Howard, B.R, Robinson, H, Sundquist, W.I, Summers, M.F, Hill, C.P. | | Deposit date: | 2007-05-22 | | Release date: | 2007-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure of the Antiviral Assembly Inhibitor CAP-1 Complex with the HIV-1 CA Protein
J.Mol.Biol., 373, 2007
|
|
5VKO
 
 | | SPT6 tSH2-RPB1 1468-1500 pT1471, pS1493 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, ISOPROPYL ALCOHOL, Transcription elongation factor SPT6 | | Authors: | Sdano, M.A, Whitby, F.G, Hill, C.P. | | Deposit date: | 2017-04-21 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A novel SH2 recognition mechanism recruits Spt6 to the doubly phosphorylated RNA polymerase II linker at sites of transcription.
Elife, 6, 2017
|
|
3PSF
 
 | |
3PSK
 
 | |
2Q71
 
 | | Uroporphyrinogen Decarboxylase G168R single mutant enzyme in complex with coproporphyrinogen-III | | Descriptor: | COPROPORPHYRINOGEN III, Uroporphyrinogen decarboxylase | | Authors: | Phillips, J.D, Whitby, F.G, Stadtmueller, B.M, Edwards, C.Q, Hill, C.P, Kushner, J.P. | | Deposit date: | 2007-06-05 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Two Novel Uropophyrinogen Decarboxylase (URO-D) Mutations Causing Hepatoerythropoietic Porphyria (HEP)
Transl.Res., 149, 2007
|
|
1FNT
 
 | | CRYSTAL STRUCTURE OF THE 20S PROTEASOME FROM YEAST IN COMPLEX WITH THE PROTEASOME ACTIVATOR PA26 FROM TRYPANOSOME BRUCEI AT 3.2 ANGSTROMS RESOLUTION | | Descriptor: | MAGNESIUM ION, PROTEASOME ACTIVATOR PROTEIN PA26, PROTEASOME COMPONENT C1, ... | | Authors: | Whitby, F.G, Masters, E, Kramer, L, Knowlton, J.R, Yao, Y, Wang, C.C, Hill, C.P. | | Deposit date: | 2000-08-23 | | Release date: | 2001-04-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the activation of 20S proteasomes by 11S regulators.
Nature, 408, 2000
|
|
7MQS
 
 | | The insulin receptor ectodomain in complex with three venom hybrid insulin molecules - asymmetric conformation | | Descriptor: | Insulin A chain, Insulin B chain, Isoform Short of Insulin receptor | | Authors: | Blakely, A.D, Xiong, X, Kim, J.H, Menting, J, Schafer, I.B, Schubert, H.L, Agrawal, R, Gutmann, T, Delaine, C, Zhang, Y, Artik, G.O, Merriman, A, Eckert, D, Lawrence, M.C, Coskun, U, Fisher, S.J, Forbes, B.E, Safavi-Hemami, H, Hill, C.P, Chou, D.H.C. | | Deposit date: | 2021-05-06 | | Release date: | 2022-03-16 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Symmetric and asymmetric receptor conformation continuum induced by a new insulin.
Nat.Chem.Biol., 18, 2022
|
|
