8WI5
 
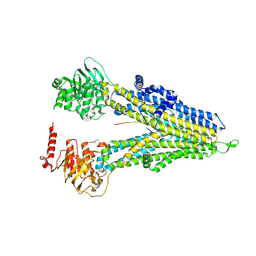 | | M5PI-bound hMRP5 | | Descriptor: | ATP-binding cassette sub-family C member 5 | | Authors: | Liu, Z.M, Huang, Y. | | Deposit date: | 2023-09-24 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Inhibition and transport mechanisms of the ABC transporter hMRP5.
Nat Commun, 15, 2024
|
|
4FYP
 
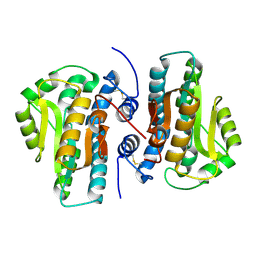 | | Crystal Structure of Plant Vegetative Storage Protein | | Descriptor: | MAGNESIUM ION, Vegetative storage protein 1 | | Authors: | Chen, Y, Wei, J, Wang, M, Gong, W, Zhang, M. | | Deposit date: | 2012-07-05 | | Release date: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of Arabidopsis VSP1 reveals the plant class C-like phosphatase structure of the DDDD superfamily of phosphohydrolases
Plos One, 7, 2012
|
|
8KCI
 
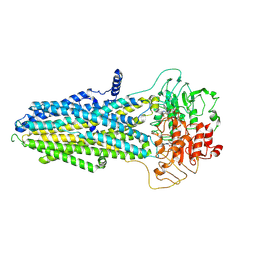 | | ATP-bound hMRP5 outward-open | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 5, MAGNESIUM ION | | Authors: | Liu, Z.M, Huang, Y. | | Deposit date: | 2023-08-07 | | Release date: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.94 Å) | | Cite: | Inhibition and transport mechanisms of the ABC transporter hMRP5.
Nat Commun, 15, 2024
|
|
7BQY
 
 | | THE CRYSTAL STRUCTURE OF COVID-19 MAIN PROTEASE IN COMPLEX WITH AN INHIBITOR N3 at 1.7 angstrom | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Liu, X, Zhang, B, Jin, Z, Yang, H, Rao, Z. | | Deposit date: | 2020-03-26 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of Mprofrom SARS-CoV-2 and discovery of its inhibitors.
Nature, 582, 2020
|
|
6LXT
 
 | | Structure of post fusion core of 2019-nCoV S2 subunit | | Descriptor: | Spike protein S2, TETRAETHYLENE GLYCOL, ZINC ION | | Authors: | Zhu, Y, Sun, F. | | Deposit date: | 2020-02-11 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Inhibition of SARS-CoV-2 (previously 2019-nCoV) infection by a highly potent pan-coronavirus fusion inhibitor targeting its spike protein that harbors a high capacity to mediate membrane fusion.
Cell Res., 30, 2020
|
|
7W1M
 
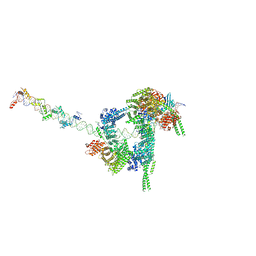 | | Cryo-EM structure of human cohesin-CTCF-DNA complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Cohesin subunit SA-1, ... | | Authors: | Shi, Z.B, Bai, X.C, Yu, H. | | Deposit date: | 2021-11-19 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | CTCF and R-loops are boundaries of cohesin-mediated DNA looping.
Mol.Cell, 83, 2023
|
|
5YF4
 
 | | A kinase complex MST4-MOB4 | | Descriptor: | MOB-like protein phocein, Peptide from Serine/threonine-protein kinase 26, ZINC ION | | Authors: | Chen, M, Zhou, Z.C. | | Deposit date: | 2017-09-20 | | Release date: | 2018-08-15 | | Last modified: | 2018-09-26 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | The MST4-MOB4 complex disrupts the MST1-MOB1 complex in the Hippo-YAP pathway and plays a pro-oncogenic role in pancreatic cancer.
J. Biol. Chem., 293, 2018
|
|
5WRO
 
 | | Crystal structure of Drosophila enolase | | Descriptor: | CADMIUM ION, CHLORIDE ION, COBALT (II) ION, ... | | Authors: | Zhang, Z, Shi, Z. | | Deposit date: | 2016-12-02 | | Release date: | 2017-04-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Crystal structure of enolase from Drosophila melanogaster.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
7X4I
 
 | |
8HEW
 
 | | Potato 14-3-3 St14f | | Descriptor: | 14-3-3 protein, StFDL1 peptide | | Authors: | Taoka, K, Kawahara, I, Shinya, S, Harada, K, Muranaka, T, Furuita, K, Nakagawa, A, Fujiwara, T, Tsuji, H, Kojima, C. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Multifunctional chemical inhibitors of the florigen activation complex discovered by structure-based high-throughput screening.
Plant J., 112, 2022
|
|
6E3E
 
 | | Structure of RORgt in complex with a novel inverse agonist. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-[(5R)-5-[(7-fluoro-1,1-dimethyl-2,3-dihydro-1H-inden-5-yl)carbamoyl]-2-methoxy-7,8-dihydro-1,6-naphthyridin-6(5H)-yl]-5-oxopentanoic acid, Nuclear receptor ROR-gamma, ... | | Authors: | Skene, R.J, Hoffman, I. | | Deposit date: | 2018-07-13 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Retinoic Acid-Related Orphan Receptor gamma t (ROR gamma t) Agonist Structure-Based Functionality Switching Approach from In House ROR gamma t Inverse Agonist to ROR gamma t Agonist.
J.Med.Chem., 62, 2019
|
|
6E3G
 
 | | Structure of RORgt in complex with a novel agonist. | | Descriptor: | (5R)-6-acetyl-2-methoxy-N-{4-[(2-methoxyphenyl)methoxy]phenyl}-5,6,7,8-tetrahydro-1,6-naphthyridine-5-carboxamide, 1,2-ETHANEDIOL, Nuclear receptor ROR-gamma, ... | | Authors: | Skene, R.J, Hoffman, I. | | Deposit date: | 2018-07-13 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Retinoic Acid-Related Orphan Receptor gamma t (ROR gamma t) Agonist Structure-Based Functionality Switching Approach from In House ROR gamma t Inverse Agonist to ROR gamma t Agonist.
J.Med.Chem., 62, 2019
|
|
7TJE
 
 | | Bacteriophage Q beta capsid protein A38K | | Descriptor: | Minor capsid protein A1 | | Authors: | Jin, X. | | Deposit date: | 2022-01-16 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Alternative Assembly of Q beta Virus-like Particles
To Be Published
|
|
7TJG
 
 | |
7TJM
 
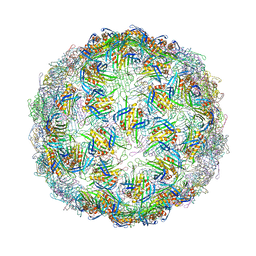 | |
7TJD
 
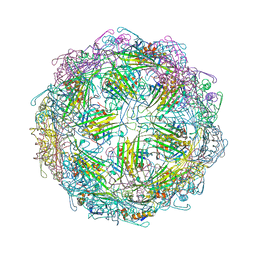 | |
3CG7
 
 | |
3CM6
 
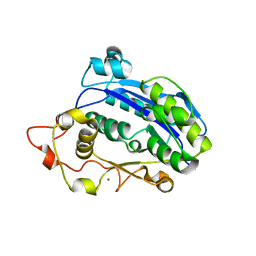 | |
4LGE
 
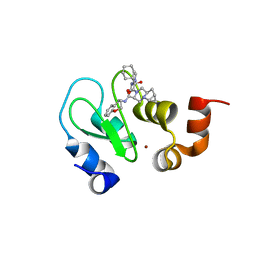 | | Crystal structure of clAP1 BIR3 bound to T3261256 | | Descriptor: | (4S,6aR,7aS)-5-{(2S)-2-cyclohexyl-2-[(N-methyl-L-alanyl)amino]acetyl}-N-[(4R)-3,4-dihydro-2H-chromen-4-yl]octahydro-1H-cyclopropa[4,5]pyrrolo[1,2-a]pyrazine-4-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Dougan, D.R. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design, stereoselective synthesis, and biological evaluation of novel tri-cyclic compounds as inhibitor of apoptosis proteins (IAP) antagonists.
Bioorg.Med.Chem., 21, 2013
|
|
4LGU
 
 | | Crystal structure of clAP1 BIR3 bound to T3226692 | | Descriptor: | (3S,8aR)-N-((R)-chroman-4-yl)-2-((S)-2-cyclohexyl-2-((S)-2-(methylamino)propanamido)acetyl)octahydropyrrolo[1,2-a]pyrazine-3-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Dougan, D.R, Mol, C.D, Snell, G.P. | | Deposit date: | 2013-06-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, stereoselective synthesis, and biological evaluation of novel tri-cyclic compounds as inhibitor of apoptosis proteins (IAP) antagonists.
Bioorg.Med.Chem., 21, 2013
|
|
4HY4
 
 | | Crystal structure of cIAP1 BIR3 bound to T3170284 | | Descriptor: | (3S,8aR)-2-{(2S)-2-cyclohexyl-2-[(N-methyl-L-alanyl)amino]acetyl}-N-[(1R)-1,2,3,4-tetrahydronaphthalen-1-yl]octahydropyrrolo[1,2-a]pyrazine-3-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Dougan, D.R, Mol, C.D, Snell, G.P. | | Deposit date: | 2012-11-12 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.249 Å) | | Cite: | Design and Synthesis of Potent Inhibitor of Apoptosis (IAP) Proteins Antagonists Bearing an Octahydropyrrolo[1,2-a]pyrazine Scaffold as a Novel Proline Mimetic.
J.Med.Chem., 56, 2013
|
|
4HY0
 
 | | Crystal structure of XIAP BIR3 with T3256336 | | Descriptor: | (3S,7R,8aR)-2-{(2S)-2-(4,4-difluorocyclohexyl)-2-[(N-methyl-L-alanyl)amino]acetyl}-N-[(4R)-3,4-dihydro-2H-chromen-4-yl]-7-ethoxyoctahydropyrrolo[1,2-a]pyrazine-3-carboxamide, E3 ubiquitin-protein ligase XIAP, ZINC ION | | Authors: | Snell, G.S, Dougan, D.R. | | Deposit date: | 2012-11-12 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Design and Synthesis of Potent Inhibitor of Apoptosis (IAP) Proteins Antagonists Bearing an Octahydropyrrolo[1,2-a]pyrazine Scaffold as a Novel Proline Mimetic.
J.Med.Chem., 56, 2013
|
|
4HY5
 
 | | Crystal structure of cIAP1 BIR3 bound to T3256336 | | Descriptor: | (3S,7R,8aR)-2-{(2S)-2-(4,4-difluorocyclohexyl)-2-[(N-methyl-L-alanyl)amino]acetyl}-N-[(4R)-3,4-dihydro-2H-chromen-4-yl]-7-ethoxyoctahydropyrrolo[1,2-a]pyrazine-3-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Dougan, D.R, Snell, G.P. | | Deposit date: | 2012-11-13 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Design and Synthesis of Potent Inhibitor of Apoptosis (IAP) Proteins Antagonists Bearing an Octahydropyrrolo[1,2-a]pyrazine Scaffold as a Novel Proline Mimetic.
J.Med.Chem., 56, 2013
|
|
9ETU
 
 | | Archaellum filament from the Halobacterium salinarum deltaAgl27 strain | | Descriptor: | 3-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-beta-D-glucopyranuronic acid-(1-4)-beta-D-glucopyranose, Archaellin | | Authors: | Grossman-Haham, I, Shahar, A. | | Deposit date: | 2024-03-27 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Perturbed N-glycosylation of Halobacterium salinarum archaellum filaments leads to filament bundling and compromised cell motility.
Nat Commun, 15, 2024
|
|
9EQ7
 
 | | Halobacterium salinarum archaellum filament | | Descriptor: | 2-O-sulfo-beta-D-glucopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-beta-D-glucopyranuronic acid-(1-4)-beta-D-glucopyranose, Archaellin | | Authors: | Grossman-Haham, I, Shahar, A. | | Deposit date: | 2024-03-21 | | Release date: | 2024-07-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Perturbed N-glycosylation of Halobacterium salinarum archaellum filaments leads to filament bundling and compromised cell motility.
Nat Commun, 15, 2024
|
|
