5K6U
 
 | | Sidekick-1 immunoglobulin domains 1-4, crystal form 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CESIUM ION, IODIDE ION, ... | | Authors: | Jin, X, Goodman, K.M, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Molecular basis of sidekick-mediated cell-cell adhesion and specificity.
Elife, 5, 2016
|
|
7TJE
 
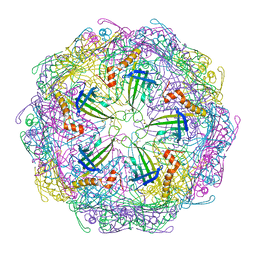 | | Bacteriophage Q beta capsid protein A38K | | Descriptor: | Minor capsid protein A1 | | Authors: | Jin, X. | | Deposit date: | 2022-01-16 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Alternative Assembly of Q beta Virus-like Particles
To Be Published
|
|
7TJM
 
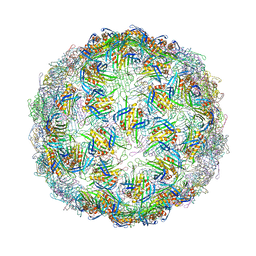 | |
7TJD
 
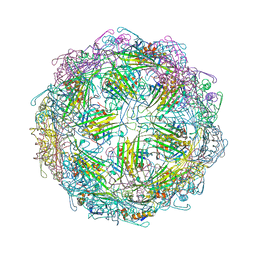 | |
7TJG
 
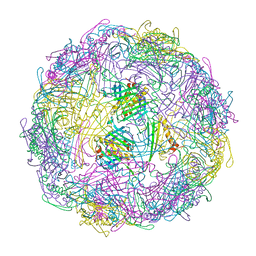 | |
1P1H
 
 | | Crystal structure of the 1L-myo-inositol/NAD+ complex | | Descriptor: | Inositol-3-phosphate synthase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Jin, X, Geiger, J.H. | | Deposit date: | 2003-04-12 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structures of NAD(+)- and NADH-bound 1-l-myo-inositol 1-phosphate synthase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1P1K
 
 | |
1P1J
 
 | | Crystal structure of the 1L-myo-inositol 1-phosphate synthase complexed with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, Inositol-3-phosphate synthase, ... | | Authors: | Jin, X, Geiger, J.H. | | Deposit date: | 2003-04-12 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of NAD(+)- and NADH-bound 1-l-myo-inositol 1-phosphate synthase.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1P1F
 
 | |
1P1I
 
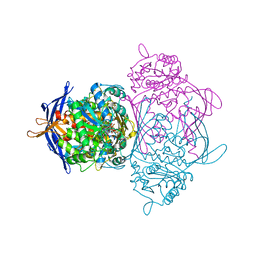 | |
1RM1
 
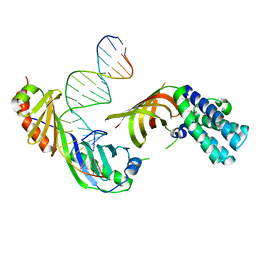 | | Structure of a Yeast TFIIA/TBP/TATA-box DNA Complex | | Descriptor: | 5'-D(*AP*TP*CP*GP*AP*TP*CP*GP*AP*TP*AP*TP*AP*AP*AP*AP*CP*G)-3', 5'-D(P*CP*GP*TP*TP*TP*TP*AP*TP*AP*TP*CP*GP*AP*TP*CP*GP*AP*T)-3', TATA-box binding protein, ... | | Authors: | Jin, X, Gewirth, D.T, Geiger, J.H. | | Deposit date: | 2003-11-26 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | High Resolution Structure of a Yeast TFIIA/TBP/TATA-box DNA Complex
TO BE PUBLISHED
|
|
1RM0
 
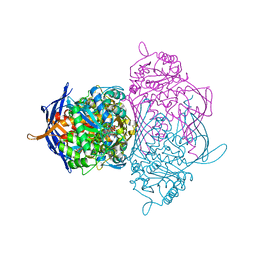 | | Crystal Structure of Myo-Inositol 1-Phosphate Synthase From Saccharomyces cerevisiae In Complex With NAD+ and 2-deoxy-D-glucitol 6-(E)-vinylhomophosphonate | | Descriptor: | (3,4,5,7-TETRAHYDROXY-HEPT-1-ENYL)-PHOSPHONIC ACID, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MANGANESE (II) ION, ... | | Authors: | Jin, X, Foley, K.M, Geiger, J.H. | | Deposit date: | 2003-11-26 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of the 1L-myo-inositol-1-phosphate synthase-NAD+-2-deoxy-D-glucitol 6-(E)-vinylhomophosphonate complex demands a revision of the enzyme mechanism.
J.Biol.Chem., 279, 2004
|
|
3BL8
 
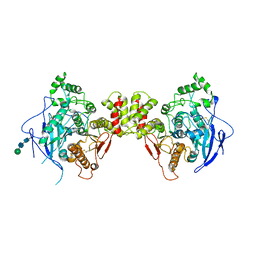 | | Crystal structure of the extracellular domain of neuroligin 2A from mouse | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neuroligin-2, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jin, X, Koehnke, J, Shapiro, L. | | Deposit date: | 2007-12-10 | | Release date: | 2008-02-19 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the extracellular cholinesterase-like domain from neuroligin-2.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1YP2
 
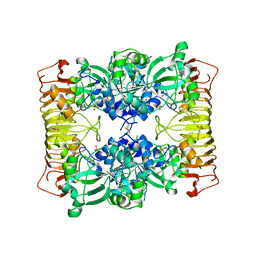 | | Crystal structure of potato tuber ADP-glucose pyrophosphorylase | | Descriptor: | Glucose-1-phosphate adenylyltransferase small subunit, PARA-MERCURY-BENZENESULFONIC ACID, SULFATE ION | | Authors: | Jin, X, Ballicora, M.A, Preiss, J, Geiger, J.H. | | Deposit date: | 2005-01-29 | | Release date: | 2005-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Crystal structure of potato tuber ADP-glucose pyrophosphorylase.
Embo J., 24, 2005
|
|
1YP3
 
 | | Crystal structure of potato tuber ADP-glucose pyrophosphorylase in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Glucose-1-phosphate adenylyltransferase small subunit, SULFATE ION | | Authors: | Jin, X, Ballicora, M.A, Preiss, J, Geiger, J.H. | | Deposit date: | 2005-01-29 | | Release date: | 2005-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of potato tuber ADP-glucose pyrophosphorylase.
Embo J., 24, 2005
|
|
1YP4
 
 | | Crystal structure of potato tuber ADP-glucose pyrophosphorylase in complex with ADP-glucose | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE-GLUCOSE, Glucose-1-phosphate adenylyltransferase small subunit, ... | | Authors: | Jin, X, Ballicora, M.A, Preiss, J, Geiger, J.H. | | Deposit date: | 2005-01-29 | | Release date: | 2005-03-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of potato tuber ADP-glucose pyrophosphorylase.
Embo J., 24, 2005
|
|
3UBF
 
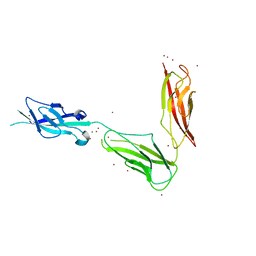 | | Crystal structure of Drosophila N-cadherin EC1-3, I | | Descriptor: | CALCIUM ION, Neural-cadherin, ZINC ION | | Authors: | Jin, X, Walker, M.A, Shapiro, L. | | Deposit date: | 2011-10-24 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Drosophila N-cadherin ectodomain regions reveal a widely used class of Ca2+-free interdomain linkers.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UBH
 
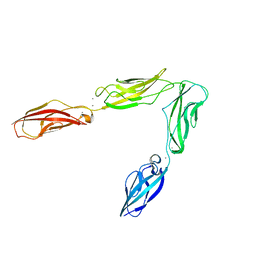 | | Crystal structure of Drosophila N-cadherin EC1-4 | | Descriptor: | CALCIUM ION, Neural-cadherin | | Authors: | Jin, X, Walker, M.A, Shapiro, L. | | Deposit date: | 2011-10-24 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of Drosophila N-cadherin ectodomain regions reveal a widely used class of Ca2+-free interdomain linkers.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3UBG
 
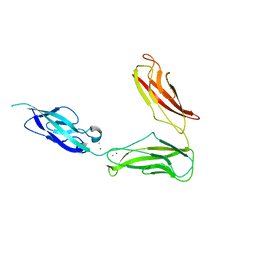 | | Crystal structure of Drosophila N-cadherin EC1-3, II | | Descriptor: | CALCIUM ION, Neural-cadherin, ZINC ION | | Authors: | Jin, X, Walker, M.A, Shapiro, L. | | Deposit date: | 2011-10-24 | | Release date: | 2011-12-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.502 Å) | | Cite: | Crystal structures of Drosophila N-cadherin ectodomain regions reveal a widely used class of Ca2+-free interdomain linkers.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3LNE
 
 | |
3LNF
 
 | |
3LND
 
 | |
3MW4
 
 | | Crystal structure of beta-neurexin 3 without the splice insert 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neurexin-2-beta, ... | | Authors: | Jin, X, Shapiro, L. | | Deposit date: | 2010-05-05 | | Release date: | 2010-07-28 | | Last modified: | 2021-10-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Splice Form Dependence of beta-Neurexin/Neuroligin Binding Interactions.
Neuron, 67, 2010
|
|
3MW3
 
 | | Crystal structure of beta-neurexin 2 with the splice insert 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neurexin-2-beta | | Authors: | Jin, X, Shapiro, L. | | Deposit date: | 2010-05-05 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Splice Form Dependence of beta-Neurexin/Neuroligin Binding Interactions.
Neuron, 67, 2010
|
|
3MW2
 
 | | Crystal structure of beta-neurexin 1 with the splice insert 4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-D-mannopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Neurexin-1-alpha, PHOSPHATE ION | | Authors: | Jin, X, Shapiro, L. | | Deposit date: | 2010-05-05 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Splice Form Dependence of beta-Neurexin/Neuroligin Binding Interactions.
Neuron, 67, 2010
|
|
