6C3N
 
 | | Crystal structure of BCL6 BTB domain in complex with compound 7CC5 | | Descriptor: | B-cell lymphoma 6 protein, N-(2-phenylethyl)-N'-pyridin-3-ylthiourea | | Authors: | Linhares, B, Cheng, H, Xue, F, Cierpicki, T. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.53170586 Å) | | Cite: | Identification of Thiourea-Based Inhibitors of the B-Cell Lymphoma 6 BTB Domain via NMR-Based Fragment Screening and Computer-Aided Drug Design.
J.Med.Chem., 61, 2018
|
|
6CQ1
 
 | | BCL6 BTB domain in complex with 15a | | Descriptor: | 2-{6-({[2-(1H-indol-3-yl)ethyl]carbamothioyl}amino)-3-[(4-methylpiperazin-1-yl)methyl]-1H-indol-1-yl}-N-(propan-2-yl)acetamide, B-cell lymphoma 6 protein | | Authors: | Linhares, B.M, Cheng, H, Xue, F, Cierpicki, T. | | Deposit date: | 2018-03-14 | | Release date: | 2019-03-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.69921041 Å) | | Cite: | Identification of Thiourea-Based Inhibitors of the B-Cell Lymphoma 6 BTB Domain via NMR-Based Fragment Screening and Computer-Aided Drug Design.
J.Med.Chem., 61, 2018
|
|
7RSJ
 
 | | Structure of the VPS34 kinase domain with compound 14 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-{4-[(7R,8R)-4-oxo-7-(propan-2-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrazin-2-yl]pyridin-2-yl}cyclopropanecarboxamide, ... | | Authors: | Hu, D.X, Patel, S, Chen, H, Wang, S, Staben, S, Dimitrova, Y.N, Wallweber, H.A, Lee, J.Y, Chan, G.K.Y, Sneeringer, C.J, Prangley, M.S, Moffat, J.G, Wu, C, Schutt, L.K, Salphati, L, Pang, J, McNamara, E, Huang, H, Chen, Y, Wang, Y, Zhao, W, Lim, J, Murthy, A, Siu, M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | Structure-Based Design of Potent, Selective, and Orally Bioavailable VPS34 Kinase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7RSP
 
 | | Structure of the VPS34 kinase domain with compound 14 | | Descriptor: | (7R,8R)-2-[(3R)-3-methylmorpholin-4-yl]-7-(propan-2-yl)-6,7-dihydropyrazolo[1,5-a]pyrazin-4(5H)-one, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3 | | Authors: | Hu, D.X, Patel, S, Chen, H, Wang, S, Staben, S, Dimitrova, Y.N, Wallweber, H.A, Lee, J.Y, Chan, G.K.Y, Sneeringer, C.J, Prangley, M.S, Moffat, J.G, Wu, C, Schutt, L.K, Salphati, L, Pang, J, McNamara, E, Huang, H, Chen, Y, Wang, Y, Zhao, W, Lim, J, Murthy, A, Siu, M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure-Based Design of Potent, Selective, and Orally Bioavailable VPS34 Kinase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7RSV
 
 | | Structure of the VPS34 kinase domain with compound 5 | | Descriptor: | (5aS,8aR,9S)-2-[(3R)-3-methylmorpholin-4-yl]-5,5a,6,7,8,8a-hexahydro-4H-cyclopenta[e]pyrazolo[1,5-a]pyrazin-4-one, GLYCEROL, Phosphatidylinositol 3-kinase catalytic subunit type 3, ... | | Authors: | Hu, D.X, Patel, S, Chen, H, Wang, S, Staben, S, Dimitrova, Y.N, Wallweber, H.A, Lee, J.Y, Chan, G.K.Y, Sneeringer, C.J, Prangley, M.S, Moffat, J.G, Wu, C, Schutt, L.K, Salphati, L, Pang, J, McNamara, E, Huang, H, Chen, Y, Wang, Y, Zhao, W, Lim, J, Murthy, A, Siu, M. | | Deposit date: | 2021-08-11 | | Release date: | 2021-11-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-Based Design of Potent, Selective, and Orally Bioavailable VPS34 Kinase Inhibitors.
J.Med.Chem., 65, 2022
|
|
6LGX
 
 | | Structure of Rabies virus glycoprotein at basic pH | | Descriptor: | Glycoprotein,Glycoprotein,Glycoprotein | | Authors: | Yang, F.L, Lin, S, Ye, F, Yang, J, Qi, J.X, Chen, Z.J, Lin, X, Wang, J.C, Yue, D, Cheng, Y.W, Chen, Z.M, Chen, H, You, Y, Zhang, Z.L, Yang, Y, Yang, M, Sun, H.L, Li, Y.H, Cao, Y, Yang, S.Y, Wei, Y.Q, Gao, G.F, Lu, G.W. | | Deposit date: | 2019-12-06 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.097 Å) | | Cite: | Structural Analysis of Rabies Virus Glycoprotein Reveals pH-Dependent Conformational Changes and Interactions with a Neutralizing Antibody.
Cell Host Microbe, 27, 2020
|
|
5CRL
 
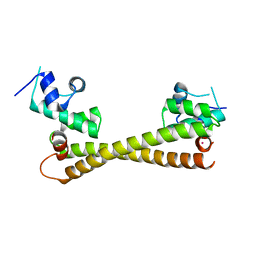 | |
8ZRY
 
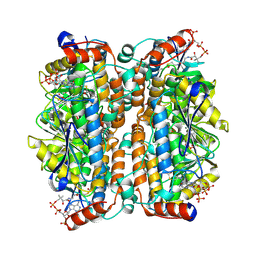 | | Structure of human ECHS1 in complex with Crotonoyl-CoA | | Descriptor: | Enoyl-CoA hydratase, mitochondrial, MAGNESIUM ION, ... | | Authors: | Su, G, Xu, Y, Chen, B, Ju, K, Sun, X, Jin, Y, Liu, D, Chen, H, Zhang, S, Luan, X. | | Deposit date: | 2024-06-05 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.23 Å) | | Cite: | Structural and biochemical mechanism of short-chain enoyl-CoA hydratase (ECHS1) substrate recognition.
Commun Biol, 8, 2025
|
|
8ZRU
 
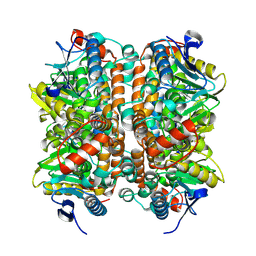 | | Structure of human ECHS1 in apo state | | Descriptor: | Enoyl-CoA hydratase, mitochondrial, MAGNESIUM ION | | Authors: | Su, G, Xu, Y, Chen, B, Ju, K, Sun, X, Jin, Y, Liu, D, Chen, H, Zhang, S, Luan, X. | | Deposit date: | 2024-06-05 | | Release date: | 2025-04-30 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.18 Å) | | Cite: | Structural and biochemical mechanism of short-chain enoyl-CoA hydratase (ECHS1) substrate recognition.
Commun Biol, 8, 2025
|
|
8ZRV
 
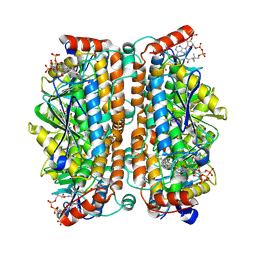 | | Structure of human ECHS1 in complex with Hexanoyl-CoA | | Descriptor: | Enoyl-CoA hydratase, mitochondrial, HEXANOYL-COENZYME A, ... | | Authors: | Su, G, Xu, Y, Chen, B, Ju, K, Sun, X, Jin, Y, Liu, D, Chen, H, Zhang, S, Luan, X. | | Deposit date: | 2024-06-05 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.55 Å) | | Cite: | Structural and biochemical mechanism of short-chain enoyl-CoA hydratase (ECHS1) substrate recognition.
Commun Biol, 8, 2025
|
|
8ZRX
 
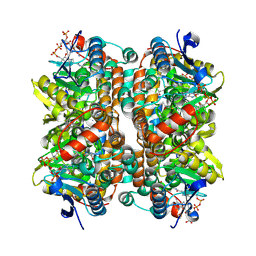 | | Structure of human ECHS1 in complex with Acetoacetyl-CoA | | Descriptor: | ACETOACETYL-COENZYME A, Enoyl-CoA hydratase, mitochondrial, ... | | Authors: | Su, G, Xu, Y, Chen, B, Ju, K, Sun, X, Jin, Y, Liu, D, Chen, H, Zhang, S, Luan, X. | | Deposit date: | 2024-06-05 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Structural and biochemical mechanism of short-chain enoyl-CoA hydratase (ECHS1) substrate recognition.
Commun Biol, 8, 2025
|
|
8ZRW
 
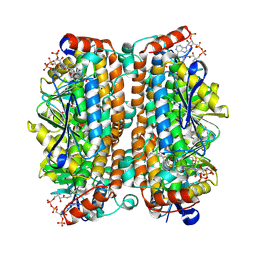 | | Structure of human ECHS1 in complex with Octanoyl-CoA | | Descriptor: | Enoyl-CoA hydratase, mitochondrial, MAGNESIUM ION, ... | | Authors: | Su, G, Xu, Y, Chen, B, Ju, K, Sun, X, Jin, Y, Liu, D, Chen, H, Zhang, S, Luan, X. | | Deposit date: | 2024-06-05 | | Release date: | 2025-04-30 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | Structural and biochemical mechanism of short-chain enoyl-CoA hydratase (ECHS1) substrate recognition.
Commun Biol, 8, 2025
|
|
8ZEQ
 
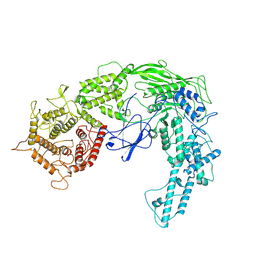 | |
8HRI
 
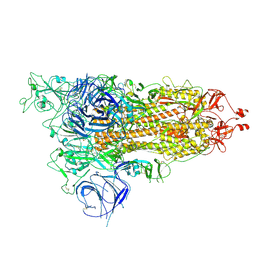 | |
8HRJ
 
 | |
6LSA
 
 | | Complex structure of bovine herpesvirus 1 glycoprotein D and bovine nectin-1 IgV | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D, ... | | Authors: | Yue, D, Chen, Z.J, Yang, F.L, Ye, F, Lin, S, Cheng, Y.W, Wang, J.C, Chen, Z.M, Lin, X, Yang, J, Chen, H, Zhang, Z.L, You, Y, Sun, H.L, Wen, A, Wang, L.L, Zheng, Y, Cao, Y, Li, Y.H, Lu, G.W. | | Deposit date: | 2020-01-17 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Crystal structure of bovine herpesvirus 1 glycoprotein D bound to nectin-1 reveals the basis for its low-affinity binding to the receptor.
Sci Adv, 6, 2020
|
|
6GCW
 
 | | Focal Adhesion Kinase catalytic domain in complex with irreversible inhibitor | | Descriptor: | 2-[[5-chloranyl-2-[[4-[[[1-[2-(propanoylamino)ethyl]-1,2,3-triazol-4-yl]methylamino]methyl]phenyl]amino]pyrimidin-4-yl]amino]-~{N}-methyl-benzamide, Focal adhesion kinase 1, SULFATE ION | | Authors: | Yen-Pon, E, Li, B, Acebron-Garcia-de-Eulate, M, Tomkiewicz-Raulet, C, Dawson, J, Lietha, D, Frame, M.C, Coumoul, X, Garbay, C, Etheve-Quelquejeu, M, Chen, H. | | Deposit date: | 2018-04-19 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design, Synthesis, and Characterization of the First Irreversible Inhibitor of Focal Adhesion Kinase.
Acs Chem.Biol., 13, 2018
|
|
6GCX
 
 | | Focal Adhesion Kinase catalytic domain in complex with irreversible inhibitor | | Descriptor: | 2-[[2-[[4-[[[3,4-bis(oxidanylidene)-2-[2-(propanoylamino)ethylamino]cyclobuten-1-yl]amino]methyl]phenyl]amino]-5-chloranyl-pyrimidin-4-yl]amino]-~{N}-methyl-benzamide, Focal adhesion kinase 1, SULFATE ION | | Authors: | Yen-Pon, E, Li, B, Acebron-Garcia de Eulate, M, Tomkiewicz-Raulet, C, Dawson, J, Lietha, D, Frame, M.C, Coumoul, X, Garbay, C, Etheve-Quelquejeu, M, Chen, H. | | Deposit date: | 2018-04-19 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.553 Å) | | Cite: | Structure-Based Design, Synthesis, and Characterization of the First Irreversible Inhibitor of Focal Adhesion Kinase.
Acs Chem.Biol., 13, 2018
|
|
6GCR
 
 | | Focal Adhesion Kinase catalytic domain in complex with irreversible inhibitor | | Descriptor: | 2-[[2-[[4-[[[3,4-bis(oxidanylidene)-2-[2-(propanoylamino)ethylamino]cyclobuten-1-yl]amino]methyl]phenyl]amino]-5-chloranyl-pyrimidin-4-yl]amino]-~{N}-methyl-benzamide, Focal adhesion kinase 1 | | Authors: | Yen-Pon, E, Li, B, Acebron-Garcia de Eulate, M, Tomkiewicz-Raulet, C, Dawson, J, Lietha, D, Frame, M.C, Coumoul, X, Garbay, C, Etheve-Quelquejeu, M, Chen, H. | | Deposit date: | 2018-04-19 | | Release date: | 2019-05-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Based Design, Synthesis, and Characterization of the First Irreversible Inhibitor of Focal Adhesion Kinase.
Acs Chem.Biol., 13, 2018
|
|
6LS9
 
 | | Crystal structure of bovine herpesvirus 1 glycoprotein D | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein D | | Authors: | Yue, D, Chen, Z.J, Yang, F.L, Ye, F, Lin, S, Cheng, Y.W, Wang, J.C, Chen, Z.M, Lin, X, Yang, J, Chen, H, Zhang, Z.L, You, Y, Sun, H.L, Wen, A, Wang, L.L, Zheng, Y, Cao, Y, Li, Y.H, Lu, G.W. | | Deposit date: | 2020-01-17 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Crystal structure of bovine herpesvirus 1 glycoprotein D bound to nectin-1 reveals the basis for its low-affinity binding to the receptor.
Sci Adv, 6, 2020
|
|
5XXE
 
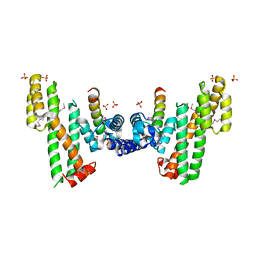 | | Crystal structure of Poz1 and Tpz1 | | Descriptor: | Protection of telomeres protein poz1, Protection of telomeres protein tpz1, SULFATE ION, ... | | Authors: | Xue, J, Chen, H, Wu, J, Lei, M. | | Deposit date: | 2017-07-03 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the fission yeast S. pombe telomeric Tpz1-Poz1-Rap1 complex.
Cell Res., 27, 2017
|
|
5XXF
 
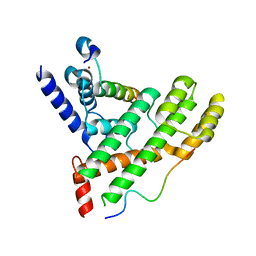 | | Crystal structure of Poz1, Tpz1 and Rap1 | | Descriptor: | Protection of telomeres protein poz1, Protection of telomeres protein tpz1, Rap1, ... | | Authors: | Xue, J, Chen, H, Wu, J, Lei, M. | | Deposit date: | 2017-07-03 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the fission yeast S. pombe telomeric Tpz1-Poz1-Rap1 complex.
Cell Res., 27, 2017
|
|
6WJ5
 
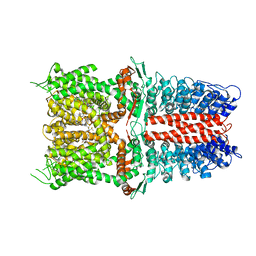 | | Structure of human TRPA1 in complex with inhibitor GDC-0334 | | Descriptor: | (4R,5S)-4-fluoro-1-[(4-fluorophenyl)sulfonyl]-5-methyl-N-({5-(trifluoromethyl)-2-[2-(trifluoromethyl)pyrimidin-5-yl]pyridin-4-yl}methyl)-L-prolinamide, Transient receptor potential cation channel subfamily A member 1 | | Authors: | Rohou, A, Rouge, L, Arthur, C.P, Volgraf, M, Chen, H. | | Deposit date: | 2020-04-11 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A TRPA1 inhibitor suppresses neurogenic inflammation and airway contraction for asthma treatment.
J.Exp.Med., 218, 2021
|
|
6LGW
 
 | | Structure of Rabies virus glycoprotein in complex with neutralizing antibody 523-11 at acidic pH | | Descriptor: | Glycoprotein, scFv 523-11 | | Authors: | Yang, F.L, Lin, S, Ye, F, Yang, J, Qi, J.X, Chen, Z.J, Lin, X, Wang, J.C, Yue, D, Cheng, Y.W, Chen, Z.M, Chen, H, You, Y, Zhang, Z.L, Yang, Y, Yang, M, Sun, H.L, Li, Y.H, Cao, Y, Yang, S.Y, Wei, Y.Q, Gao, G.F, Lu, G.W. | | Deposit date: | 2019-12-06 | | Release date: | 2020-02-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.9037 Å) | | Cite: | Structural Analysis of Rabies Virus Glycoprotein Reveals pH-Dependent Conformational Changes and Interactions with a Neutralizing Antibody.
Cell Host Microbe, 27, 2020
|
|
6KZ5
 
 | | Crystal Structure Analysis of the Csn-B-bounded NUR77 Ligand binding Domain | | Descriptor: | Nuclear receptor subfamily 4 group A member 1, ethyl 2-[2-octanoyl-3,5-bis(oxidanyl)phenyl]ethanoate | | Authors: | Hong, W, Chen, H, Wu, Q, Lin, T. | | Deposit date: | 2019-09-23 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.45 Å) | | Cite: | Blocking PPAR gamma interaction facilitates Nur77 interdiction of fatty acid uptake and suppresses breast cancer progression.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
