3D2Z
 
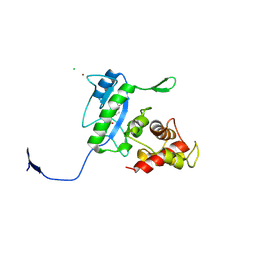 | | Complex of the N-acetylmuramyl-L-alanine amidase AmiD from E.coli with the product L-Ala-D-gamma-Glu-L-Lys | | Descriptor: | CHLORIDE ION, L-Ala-D-gamma-Glu-L-Lys peptide, N-acetylmuramoyl-L-alanine amidase amiD, ... | | Authors: | Kerff, F, Petrella, S, Herman, R, Sauvage, E, Mercier, F, Luxen, A, Frere, J.M, Joris, B, Charlier, P. | | Deposit date: | 2008-05-09 | | Release date: | 2009-06-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Specific Structural Features of the N-Acetylmuramoyl-l-Alanine Amidase AmiD from Escherichia coli and Mechanistic Implications for Enzymes of This Family.
J.Mol.Biol., 397, 2010
|
|
3TEI
 
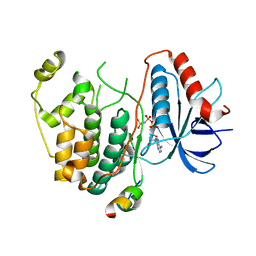 | | Crystal structure of human ERK2 complexed with a MAPK docking peptide | | Descriptor: | Mitogen-activated protein kinase 1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Ribosomal protein S6 kinase alpha-1 | | Authors: | Gogl, G, Remenyi, A. | | Deposit date: | 2011-08-15 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.404 Å) | | Cite: | Specificity of linear motifs that bind to a common mitogen-activated protein kinase docking groove.
Sci.Signal., 5, 2012
|
|
4JAJ
 
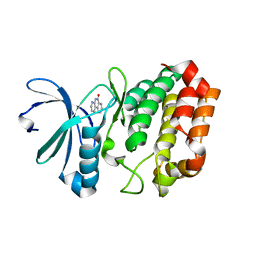 | | Crystal Structure of Aurora Kinase A in complex with BENZO[C][1,8]NAPHTHYRIDIN-6(5H)-ONE | | Descriptor: | Aurora kinase A, benzo[c][1,8]naphthyridin-6(5H)-one | | Authors: | Jiang, X, Josephson, K, Huck, B, Goutopoulos, A, Karra, S. | | Deposit date: | 2013-02-18 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SAR and evaluation of novel 5H-benzo[c][1,8]naphthyridin-6-one analogs as Aurora kinase inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
1Q9B
 
 | |
3D2Y
 
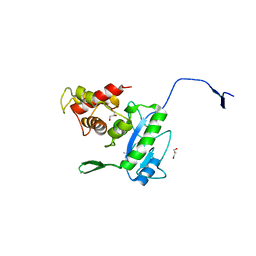 | | Complex of the N-acetylmuramyl-L-alanine amidase AmiD from E.coli with the substrate anhydro-N-acetylmuramic acid-L-Ala-D-gamma-Glu-L-Lys | | Descriptor: | Anhydro-N-acetylmuramic acid-L-Ala-D-gamma-Glu-L-Lys, GLYCEROL, N-acetylmuramoyl-L-alanine amidase amiD | | Authors: | Kerff, F, Petrella, S, Herman, R, Sauvage, E, Mercier, F, Luxen, A, Frere, J.M, Joris, B, Charlier, P. | | Deposit date: | 2008-05-09 | | Release date: | 2009-06-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Specific Structural Features of the N-Acetylmuramoyl-l-Alanine Amidase AmiD from Escherichia coli and Mechanistic Implications for Enzymes of This Family.
J.Mol.Biol., 397, 2010
|
|
2GHI
 
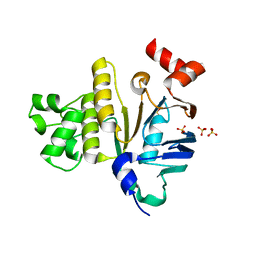 | | Crystal Structure of Plasmodium yoelii Multidrug Resistance Protein 2 | | Descriptor: | SULFATE ION, transport protein | | Authors: | Dong, A, Gao, M, Choe, J, Zhao, Y, Lew, J, Wasney, G, Alam, Z, Melone, M, Kozieradzki, I, Vedadi, M, Edwards, A.M, Arrowsmith, C.H, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-03-27 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
3QGM
 
 | | p-nitrophenyl phosphatase from Archaeoglobus fulgidus | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, p-nitrophenyl phosphatase (Pho2) | | Authors: | Osipiuk, J, Zheng, H, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-01-24 | | Release date: | 2011-02-09 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | p-nitrophenyl phosphatase from Archaeoglobus fulgidus.
To be Published
|
|
1Q8B
 
 | |
3HAE
 
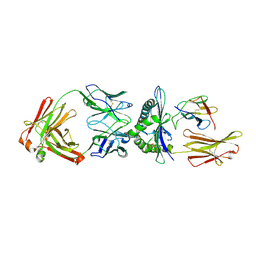 | | Rational development of high-affinity T-cell receptor-like antibodies | | Descriptor: | Antibody heavy chain, Antibody light chain, Beta-2-microglobulin, ... | | Authors: | Stewart-Jones, G, Wadle, A, Hombach, A, Shenderov, E, Held, G, Fischer, E. | | Deposit date: | 2009-05-01 | | Release date: | 2009-05-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Rational development of high-affinity T-cell receptor-like antibodies
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2JQ5
 
 | | Solution structure of RPA3114, a SEC-C motif containing protein from Rhodopseudomonas palustris; Northeast Structural Genomics Consortium target RpT5 / Ontario Center for Structural Proteomics target RP3097 | | Descriptor: | SEC-C motif, ZINC ION | | Authors: | Lemak, A, Lukin, J, Yee, A, Gutmanas, A, Karra, M, Semesi, A, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2007-05-29 | | Release date: | 2007-06-26 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a Se-C motif containing protein from Rhodopseudomonas palustris
To be Published
|
|
1R0U
 
 | | Crystal structure of ywiB protein from Bacillus subtilis | | Descriptor: | GLYCEROL, protein ywiB | | Authors: | Osipiuk, J, Xu, X, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-09-23 | | Release date: | 2003-12-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of ywiB protein from Bacillus subtilis
to be published
|
|
2R6O
 
 | | Crystal structure of putative diguanylate cyclase/phosphodiesterase from Thiobacillus denitrificans | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Putative diguanylate cyclase/phosphodiesterase (GGDEF & EAL domains) | | Authors: | Chang, C, Xu, X, Zheng, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-09-06 | | Release date: | 2007-09-18 | | Last modified: | 2012-10-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into the mechanism of c-di-GMP hydrolysis by EAL domain phosphodiesterases.
J.Mol.Biol., 402, 2010
|
|
4BCW
 
 | | Carbonic anhydrase IX mimic in complex with (E)-2-(5-bromo-2- hydroxyphenyl)ethenesulfonic acid | | Descriptor: | (E)-2-(5-BROMO-2-HYDROXYPHENYL)ETHENESULFONIC ACID, CARBONIC ANHYDRASE 2, ZINC ION | | Authors: | Tars, K, Leitans, J, Kazaks, A. | | Deposit date: | 2012-10-03 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Sulfocoumarins (1,2-Benzoxathiine-2,2-Dioxides): A Class of Potent and Isoform-Selective Inhibitors of Tumor-Associated Carbonic Anhydrases.
J.Med.Chem., 56, 2013
|
|
1EKO
 
 | | PIG ALDOSE REDUCTASE COMPLEXED WITH IDD384 INHIBITOR | | Descriptor: | ALDOSE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [2,6-DIMETHYL-4-(2-O-TOLYL-ACETYLAMINO)-BENZENESULFONYL]-GLYCINE | | Authors: | Podjarny, A. | | Deposit date: | 2000-03-09 | | Release date: | 2000-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of human aldose reductase bound to the inhibitor IDD384.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
4JZY
 
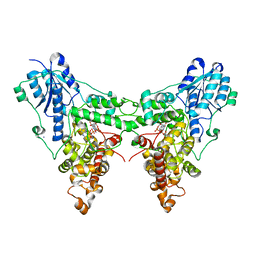 | | Crystal structures of Drosophila Cryptochrome | | Descriptor: | AMMONIUM ION, Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Czarna, A, Wolf, E. | | Deposit date: | 2013-04-03 | | Release date: | 2013-06-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function.
Cell(Cambridge,Mass.), 153, 2013
|
|
4K03
 
 | | Crystal structure of Drosophila Cryprochrome | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Berndt, A, Wolf, E. | | Deposit date: | 2013-04-03 | | Release date: | 2013-06-26 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of Drosophila cryptochrome and mouse cryptochrome1 provide insight into circadian function.
Cell(Cambridge,Mass.), 153, 2013
|
|
2L2D
 
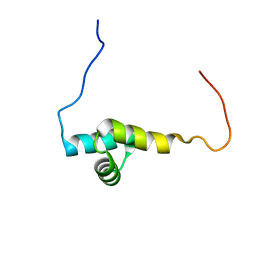 | | Solution NMR Structure of human UBA-like domain of OTUD7A_11_83, NESG target HT6304A/OCSP target OTUD7A_11_83/SGC-Toronto | | Descriptor: | OTU domain-containing protein 7A | | Authors: | Wu, B, Yee, A, Lemak, A, Gutmanas, A, Houliston, S, Semesi, A, Dhe-Paganon, S, Montelione, G.T, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP), Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-17 | | Release date: | 2010-09-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The amino-terminal UBA domain of OTUD7A
To be Published
|
|
1EQD
 
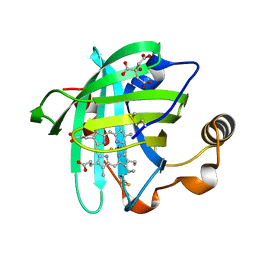 | | CRYSTAL STRUCTURE OF NITROPHORIN 4 COMPLEXED WITH CN | | Descriptor: | 5,8-DIMETHYL-1,2,3,4-TETRAVINYLPORPHINE-6,7-DIPROPIONIC ACID FERROUS COMPLEX, CITRIC ACID, CYANIDE ION, ... | | Authors: | Weichsel, A, Andersen, J.F, Roberts, S.A, Montfort, W.R. | | Deposit date: | 2000-04-03 | | Release date: | 2000-05-03 | | Last modified: | 2018-04-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nitric oxide binding to nitrophorin 4 induces complete distal pocket burial.
Nat.Struct.Biol., 7, 2000
|
|
2LXX
 
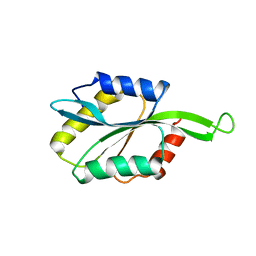 | | Solution structure of cofilin like UNC-60B protein from Caenorhabditis elegans | | Descriptor: | Actin-depolymerizing factor 2, isoform c | | Authors: | Shukla, V, Yadav, R, Kabra, A, Jain, A, Kumar, D, Ono, S, Arora, A. | | Deposit date: | 2012-09-04 | | Release date: | 2013-10-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure and dynamics of UNC-60B from Caenorhabditis elegans
To be Published
|
|
1ES6
 
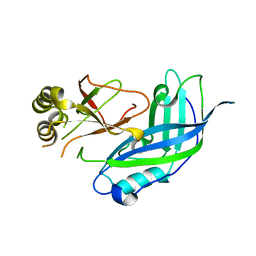 | | CRYSTAL STRUCTURE OF THE MATRIX PROTEIN OF EBOLA VIRUS | | Descriptor: | MATRIX PROTEIN VP40 | | Authors: | Dessen, A, Volchkov, V, Dolnik, O, Klenk, H.-D, Weissenhorn, W. | | Deposit date: | 2000-04-07 | | Release date: | 2000-08-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the matrix protein VP40 from Ebola virus.
EMBO J., 19, 2000
|
|
3ICF
 
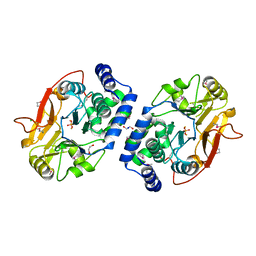 | | Structure of Protein serine/threonine phosphatase from Saccharomyces cerevisiae with similarity to human phosphatase PP5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Singer, A.U, Xu, X, Chang, C, Cui, H, Kagan, O, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of Protein serine/threonine phosphatase from Saccharomyces cerevisiae with similarity to human phosphatase PP5
To be Published
|
|
5AD3
 
 | | Bivalent binding to BET bromodomains | | Descriptor: | 3-methoxy-N-[2-[4-[1-(3-methoxy-[1,2,4]triazolo[4,3-b]pyridazin-6-yl)-4-piperidyl]phenoxy]ethyl]-N-methyl-[1,2,4]triazolo[4,3-b]pyridazin-6-amine, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Waring, M.J, Chen, H, Rabow, A.A, Walker, G, Bobby, R, Boiko, S, Bradbury, R.H, Callis, R, Dale, I, Daniels, D, Flavell, L, Holdgate, G, Jowitt, T.A, Kikhney, A, McAlister, M, Ogg, D, Patel, J, Petteruti, P, Robb, G.R, Robers, M, Stratton, N, Svergun, D.I, Wang, W, Whittaker, D. | | Deposit date: | 2015-08-19 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Potent and Selective Bivalent Inhibitors of Bet Bromodomains
Nat.Chem.Biol., 12, 2016
|
|
2CSB
 
 | | Crystal structure of Topoisomerase V from Methanopyrus kandleri (61 kDa fragment) | | Descriptor: | MAGNESIUM ION, Topoisomerase V | | Authors: | Taneja, B, Patel, A, Slesarev, A, Mondragon, A. | | Deposit date: | 2005-05-21 | | Release date: | 2006-01-31 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the N-terminal fragment of topoisomerase V reveals a new family of topoisomerases
Embo J., 25, 2006
|
|
2M8D
 
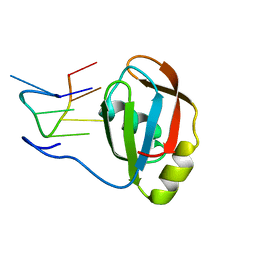 | | Structure of SRSF1 RRM2 in complex with the RNA 5'-UGAAGGAC-3' | | Descriptor: | RNA (5'-R(*UP*GP*AP*AP*GP*GP*AP*C)-3'), Serine/arginine-rich splicing factor 1 | | Authors: | Clery, A, Sinha, R, Anczukow, O, Corrionero, A, Moursy, A, Daubner, G, Valcarcel, J, Krainer, A.R, Allain, F.H.T. | | Deposit date: | 2013-05-17 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Isolated pseudo-RNA-recognition motifs of SR proteins can regulate splicing using a noncanonical mode of RNA recognition.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6R1G
 
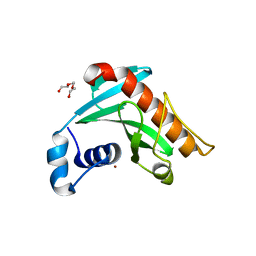 | | Crystal structure of Borrelia burgdorferi periplasmic protein BB0365 (IPLA7, p22) | | Descriptor: | Outer surface 22 kDa lipoprotein, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Brangulis, K, Akopjana, I, Petrovskis, I, Kazaks, A, Tars, K. | | Deposit date: | 2019-03-14 | | Release date: | 2019-09-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis of Borrelia burgdorferi periplasmic lipoprotein BB0365 involved in Lyme disease infection.
Febs Lett., 594, 2020
|
|
