3VM7
 
 | | Structure of an Alpha-Amylase from Malbranchea cinnamomea | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, ... | | Authors: | Zhou, P, Hu, S.Q, Zhou, Y, Han, P, Yang, S.Q, Jiang, Z.Q. | | Deposit date: | 2011-12-09 | | Release date: | 2013-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | A Novel Multifunctional alpha-Amylase from the Thermophilic Fungus Malbranchea cinnamomea: Biochemical Characterization and Three-Dimensional Structure.
Appl Biochem Biotechnol., 170, 2013
|
|
7FEE
 
 | | Crystal structure of the allosteric modulator ZCZ011 binding to CP55940-bound cannabinoid receptor 1 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-[(1R,2R,5R)-5-hydroxy-2-(3-hydroxypropyl)cyclohexyl]-5-(2-methyloctan-2-yl)phenol, 6-methyl-3-[(1S)-2-nitro-1-thiophen-2-yl-ethyl]-2-phenyl-1H-indole, ... | | Authors: | Wang, X, Zhao, C, Shao, Z. | | Deposit date: | 2021-07-19 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular mechanism of allosteric modulation for the cannabinoid receptor CB1.
Nat.Chem.Biol., 18, 2022
|
|
8HIT
 
 | | Crystal structure of anti-CTLA-4 humanized IgG1 MAb--JS007 in complex with human CTLA-4 | | Descriptor: | Cytotoxic T-lymphocyte protein 4, JS007-VH, JS007-VL | | Authors: | Tan, S, Shi, Y, Wang, Q, Gao, G.F, Guan, J, Chai, Y, Qi, J. | | Deposit date: | 2022-11-21 | | Release date: | 2023-02-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Characterization of the high-affinity anti-CTLA-4 monoclonal antibody JS007 for immune checkpoint therapy of cancer.
Mabs, 15, 2023
|
|
8HGW
 
 | | Crystal structure of MehpH in complex with MBP | | Descriptor: | 1-BUTANOL, Monoalkyl phthalate hydrolase, PHTHALIC ACID | | Authors: | Zhang, Z.M, Wang, Y.J, Chen, Y.B. | | Deposit date: | 2022-11-15 | | Release date: | 2023-03-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.80001163 Å) | | Cite: | Molecular insights into the catalytic mechanism of plasticizer degradation by a monoalkyl phthalate hydrolase.
Commun Chem, 6, 2023
|
|
8HGV
 
 | |
7C88
 
 | | Complex structure of JS003 and PD-L1 | | Descriptor: | JS003 Heavy chain, JS003 Light chain, Programmed cell death 1 ligand 1 | | Authors: | Bi, X, Shi, R, Chai, Y, Qi, J, Yan, J, Tan, S. | | Deposit date: | 2020-05-29 | | Release date: | 2021-04-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Identification of a hotspot on PD-L1 for pH-dependent binding by monoclonal antibodies for tumor therapy.
Signal Transduct Target Ther, 5, 2020
|
|
8I71
 
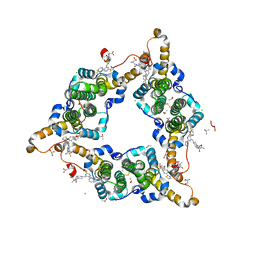 | | Hepatitis B virus core protein Y132A mutant in complex with Linvencorvir (RG7907), a Hepatitis B Virus (HBV) Core Protein Allosteric Modulator (CpAM) | | Descriptor: | 3-[(8~{a}~{S})-7-[[5-ethoxycarbonyl-4-(3-fluoranyl-2-methyl-phenyl)-2-(1,3-thiazol-2-yl)-1,4-dihydropyrimidin-6-yl]methyl]-3-oxidanylidene-5,6,8,8~{a}-tetrahydro-1~{H}-imidazo[1,5-a]pyrazin-2-yl]-2,2-dimethyl-propanoic acid, CHLORIDE ION, Capsid protein, ... | | Authors: | Zhou, Z, Xu, Z.H. | | Deposit date: | 2023-01-30 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Linvencorvir (RG7907), a Hepatitis B Virus Core Protein Allosteric Modulator, for the Treatment of Chronic HBV Infection.
J.Med.Chem., 66, 2023
|
|
2M9L
 
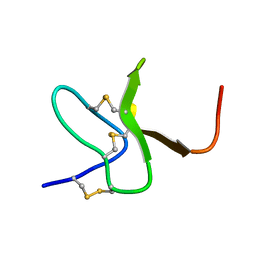 | | Solution structure of protoxin-1 | | Descriptor: | Beta-theraphotoxin-Tp1a | | Authors: | Daly, N. | | Deposit date: | 2013-06-13 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | A tarantula-venom peptide antagonizes the TRPA1 nociceptor ion channel by binding to the S1-S4 gating domain.
Curr.Biol., 24, 2014
|
|
7X8X
 
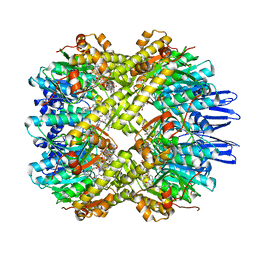 | | structural insights into Mycobacterium tuberculosis ClpP1P2 inhibition by Cediranib: implications for developing antimicrobial agents targeting Clp protease | | Descriptor: | 4-[1-[3-[4-[(4-fluoranyl-2-methyl-1H-indol-5-yl)oxy]-6-methoxy-quinazolin-7-yl]oxypropyl]piperidin-4-yl]benzamide, 4-[[3,5-bis(fluoranyl)phenyl]methyl]-N-[(4-bromophenyl)methyl]piperazine-1-carboxamide, ATP-dependent Clp protease proteolytic subunit 1, ... | | Authors: | Bao, R, Luo, Y.F, Zhu, Y.B, Yang, Y, Zhou, Y.Z. | | Deposit date: | 2022-03-15 | | Release date: | 2023-04-12 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Discovery and Mechanistic Study of Novel Mycobacterium tuberculosis ClpP1P2 Inhibitors
J.Med.Chem., 66, 2023
|
|
4Y7N
 
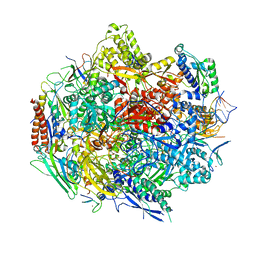 | | The Structure Insight into 5-Carboxycytosine Recognition by RNA Polymerase II during Transcription Elongation. | | Descriptor: | DNA (29-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2015-02-15 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex.
Nature, 523, 2015
|
|
4Y52
 
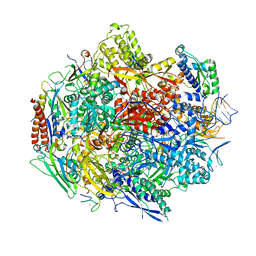 | | Crystal structure of 5-Carboxycytosine Recognition by RNA Polymerase II during Transcription Elongation. | | Descriptor: | DNA (29-MER), DNA (5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Wang, L, Chong, J, Wang, D. | | Deposit date: | 2015-02-11 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular basis for 5-carboxycytosine recognition by RNA polymerase II elongation complex.
Nature, 523, 2015
|
|
8HGH
 
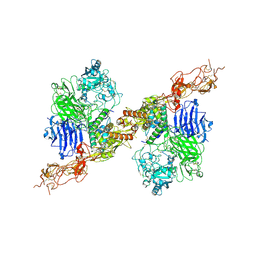 | | Structure of 2:2 PAPP-A.STC2 complex | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Pappalysin-1, Stanniocalcin-2, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
7VF6
 
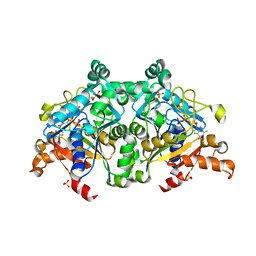 | | The crystal structure of PurZ0 | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Tong, Y, Zhang, Y. | | Deposit date: | 2021-09-10 | | Release date: | 2023-05-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | Alternative Z-genome biosynthesis pathway shows evolutionary progression from Archaea to phage.
Nat Microbiol, 8, 2023
|
|
8HGG
 
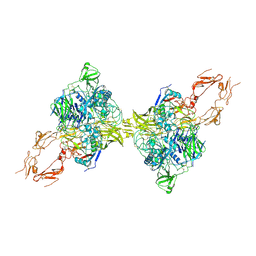 | | Structure of 2:2 PAPP-A.ProMBP complex | | Descriptor: | Bone marrow proteoglycan, Pappalysin-1, ZINC ION | | Authors: | Zhong, Q.H, Chu, H.L, Wang, G.P, Zhang, C, Wei, Y, Qiao, J, Hang, J. | | Deposit date: | 2022-11-14 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Structural insights into the covalent regulation of PAPP-A activity by proMBP and STC2.
Cell Discov, 8, 2022
|
|
4NML
 
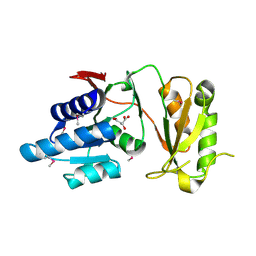 | | 2.60 Angstrom resolution crystal structure of putative ribose 5-phosphate isomerase from Toxoplasma gondii ME49 in complex with DL-Malic acid | | Descriptor: | CHLORIDE ION, D-MALATE, Ribulose 5-phosphate isomerase | | Authors: | Halavaty, A.S, Dubrovska, I, Flores, K, Shanmugam, D, Shuvalova, L, Roos, D, Ruan, J, Ngo, H, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-15 | | Release date: | 2013-12-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
8H2X
 
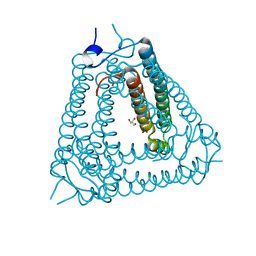 | | Structure of Acb2 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, p26 | | Authors: | Feng, Y, Cao, X.L. | | Deposit date: | 2022-10-07 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Bacteriophages inhibit and evade cGAS-like immune function in bacteria.
Cell, 186, 2023
|
|
8H2J
 
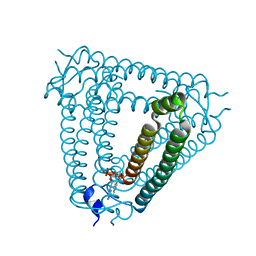 | | Structure of Acb2 complexed with 3',3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, p26 | | Authors: | Feng, Y, Cao, X.L. | | Deposit date: | 2022-10-06 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Bacteriophages inhibit and evade cGAS-like immune function in bacteria.
Cell, 186, 2023
|
|
8H39
 
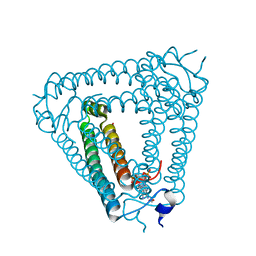 | | Structure of Acb2 complexed with c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, 1,2-ETHANEDIOL, p26 | | Authors: | Feng, Y, Cao, X.L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-02-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Bacteriophages inhibit and evade cGAS-like immune function in bacteria.
Cell, 186, 2023
|
|
4NOG
 
 | | Crystal structure of a putative ornithine aminotransferase from Toxoplasma gondii ME49 in complex with pyrodoxal-5'-phosphate | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, BETA-MERCAPTOETHANOL, ... | | Authors: | Filippova, E.V, Halavaty, A, Ruan, J, Shuvalova, L, Flores, K, Dubrovska, I, Ngo, H, Shanmugam, D, Roos, D, Anderson, W.F, Center for Structural Genomics of Infectious Diseases, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-11-19 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
4NU7
 
 | | 2.05 Angstrom Crystal Structure of Ribulose-phosphate 3-epimerase from Toxoplasma gondii. | | Descriptor: | CHLORIDE ION, Ribulose-phosphate 3-epimerase, SULFATE ION, ... | | Authors: | Minasov, G, Ruan, J, Ngo, H, Shuvalova, L, Dubrovska, I, Flores, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-12-03 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | CSGID Solves Structures and Identifies Phenotypes for Five Enzymes in Toxoplasma gondii .
Front Cell Infect Microbiol, 8, 2018
|
|
7WEE
 
 | |
7WEB
 
 | |
7WE9
 
 | | SARS-CoV-2 Omicron variant spike protein in complex with Fab XGv289 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2021-12-23 | | Release date: | 2022-05-04 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Memory B cell repertoire from triple vaccinees against diverse SARS-CoV-2 variants.
Nature, 603, 2022
|
|
7XAT
 
 | | Structure of somatostatin receptor 2 bound with SST14. | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Bo, Q, Yang, F, Li, Y.G, Meng, X.Y, Zhang, H.H, Zhou, Y.X, Ling, S.L, Sun, D.M, Lv, P, Liu, L, Shi, P, Tian, C.L. | | Deposit date: | 2022-03-19 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural insights into the activation of somatostatin receptor 2 by cyclic SST analogues.
Cell Discov, 8, 2022
|
|
7XAU
 
 | | Structure of somatostatin receptor 2 bound with octreotide. | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Bo, Q, Yang, F, Li, Y.G, Meng, X.Y, Zhang, H.H, Zhou, Y.X, Ling, S.L, Sun, D.M, Lv, P, Liu, L, Shi, P, Tian, C.L. | | Deposit date: | 2022-03-19 | | Release date: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structural insights into the activation of somatostatin receptor 2 by cyclic SST analogues.
Cell Discov, 8, 2022
|
|
