8E56
 
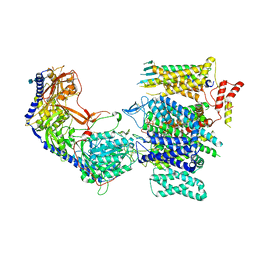 | | Rabbit L-type voltage-gated calcium channel Cav1.1 in the presence of Amiodarone at 2.8 Angstrom resolution | | Descriptor: | (2-butyl-1-benzofuran-3-yl){4-[2-(diethylamino)ethoxy]-3,5-diiodophenyl}methanone, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-08-20 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis for the severe adverse interaction of sofosbuvir and amiodarone on L-type Ca v channels.
Cell, 185, 2022
|
|
8E58
 
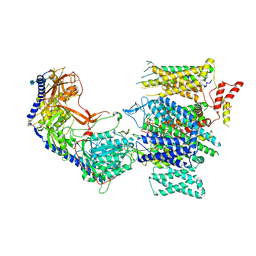 | | Rabbit L-type voltage-gated calcium channel Cav1.1 in the presence of Amiodarone and 1 mM MNI-1 at 3.0 Angstrom resolution | | Descriptor: | (2-butyl-1-benzofuran-3-yl){4-[2-(diethylamino)ethoxy]-3,5-diiodophenyl}methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2022-08-20 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis for the severe adverse interaction of sofosbuvir and amiodarone on L-type Ca v channels.
Cell, 185, 2022
|
|
7S5H
 
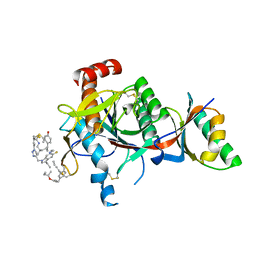 | | PCSK9(deltaCRD) in complex with cyclic peptide 35 | | Descriptor: | (2E)-but-2-ene-1,4-diol, Pro-peptide from Proprotein convertase subtilisin/kexin type 9, Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Orth, P. | | Deposit date: | 2021-09-10 | | Release date: | 2021-11-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.272 Å) | | Cite: | A Series of Novel, Highly Potent, and Orally Bioavailable Next-Generation Tricyclic Peptide PCSK9 Inhibitors.
J.Med.Chem., 64, 2021
|
|
6JKK
 
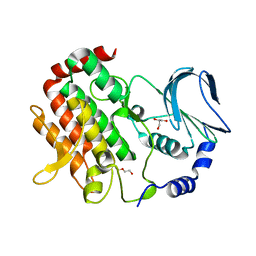 | | Crystal structure of BubR1 kinase domain | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Mitotic checkpoint control protein kinase BUB1 | | Authors: | Lin, L, Ye, S, Huang, Y, Liu, X, Zhang, R, Yao, X. | | Deposit date: | 2019-03-01 | | Release date: | 2019-06-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | BubR1 phosphorylates CENP-E as a switch enabling the transition from lateral association to end-on capture of spindle microtubules.
Cell Res., 29, 2019
|
|
8Y4U
 
 | | Crystal structure of a His1 from oryza sativa | | Descriptor: | FE (III) ION, Fe(II)/2-oxoglutarate-dependent oxygenase | | Authors: | Wang, N, Ma, J.M, Shibing, H, Beibei, Y, He, Z, Dandan, L. | | Deposit date: | 2024-01-30 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of HPPD inhibitor sensitive protein from Oryza sativa.
Biochem.Biophys.Res.Commun., 704, 2024
|
|
7S5G
 
 | | PCSK9 in complex with compound 19 | | Descriptor: | (2E)-but-2-ene-1,4-diol, GLYCEROL, Propeptide of Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Orth, P. | | Deposit date: | 2021-09-10 | | Release date: | 2021-11-03 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.041 Å) | | Cite: | A Series of Novel, Highly Potent, and Orally Bioavailable Next-Generation Tricyclic Peptide PCSK9 Inhibitors.
J.Med.Chem., 64, 2021
|
|
8YRH
 
 | | Complex of SARS-CoV-2 main protease and Rosmarinic acid | | Descriptor: | (2R)-3-(3,4-dihydroxyphenyl)-2-{[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]oxy}propanoic acid, 3C-like proteinase nsp5 | | Authors: | Wang, Q.S, Li, Q.H. | | Deposit date: | 2024-03-21 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.841 Å) | | Cite: | Structural basis of rosmarinic acid inhibitory mechanism on SARS-CoV-2 main protease.
Biochem.Biophys.Res.Commun., 724, 2024
|
|
8H3T
 
 | | The crystal structure of AlpH | | Descriptor: | AlpH, GLYCEROL | | Authors: | Zhao, Y, Li, M, Jiang, M, Pan, L.F. | | Deposit date: | 2022-10-09 | | Release date: | 2023-09-13 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.866 Å) | | Cite: | O-methyltransferase-like enzyme catalyzed diazo installation in polyketide biosynthesis.
Nat Commun, 14, 2023
|
|
4GEH
 
 | |
3BC5
 
 | | X-ray crystal structure of human ppar gamma with 2-(5-(3-(2-(5-methyl-2-phenyloxazol-4-yl)ethoxy)benzyl)-2-phenyl-2h-1,2,3-triazol-4-yl)acetic acid | | Descriptor: | (5-{3-[2-(5-methyl-2-phenyl-1,3-oxazol-4-yl)ethoxy]benzyl}-2-phenyl-2H-1,2,3-triazol-4-yl)acetic acid, Peroxisome proliferator-activated receptor gamma | | Authors: | Muckelbauer, J.K. | | Deposit date: | 2007-11-12 | | Release date: | 2008-11-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Design, synthesis and structure-activity relationships of azole acids as novel, potent dual PPAR alpha/gamma agonists.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
8TGD
 
 | | STX-478, a Mutant-Selective, Allosteric Inhibitor bound to H1047R PI3Kalpha | | Descriptor: | 1,2-ETHANEDIOL, N-(2-aminopyrimidin-5-yl)-N'-[(1R)-1-(5,7-difluoro-3-methyl-1-benzofuran-2-yl)-2,2,2-trifluoroethyl]urea, N~2~-{(4S,11aP)-2-[(4S)-4-(difluoromethyl)-2-oxo-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-L-alaninamide, ... | | Authors: | Hilbert, B, Brooijmans, N, Buckbinder, L, St.Jean Jr, D.J. | | Deposit date: | 2023-07-12 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.928 Å) | | Cite: | STX-478, a Mutant-Selective, Allosteric PI3K alpha Inhibitor Spares Metabolic Dysfunction and Improves Therapeutic Response in PI3K alpha-Mutant Xenografts.
Cancer Discov, 13, 2023
|
|
8TDU
 
 | | STX-478, a Mutant-Selective, Allosteric Inhibitor bound to PI3Kalpha | | Descriptor: | N-(2-aminopyrimidin-5-yl)-N'-[(1R)-1-(5,7-difluoro-3-methyl-1-benzofuran-2-yl)-2,2,2-trifluoroethyl]urea, N~2~-{(4S,11aP)-2-[(4S)-4-(difluoromethyl)-2-oxo-1,3-oxazolidin-3-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepin-9-yl}-L-alaninamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, ... | | Authors: | Hilbert, B.J, Brooijmans, N, Buckbinder, L, St.Jean Jr, D.J. | | Deposit date: | 2023-07-05 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | STX-478, a Mutant-Selective, Allosteric PI3K alpha Inhibitor Spares Metabolic Dysfunction and Improves Therapeutic Response in PI3K alpha-Mutant Xenografts.
Cancer Discov, 13, 2023
|
|
7DEU
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody scFv | | Authors: | Zhang, Z, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
8TNO
 
 | |
7DEO
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Spike protein S1, ... | | Authors: | Fu, D, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-04 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
7DET
 
 | | Crystal structure of SARS-CoV-2 RBD in complex with a neutralizing antibody scFv | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody scFv | | Authors: | Wang, Y, Zhang, G, Li, X, Rao, Z, Guo, Y. | | Deposit date: | 2020-11-05 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for SARS-CoV-2 neutralizing antibodies with novel binding epitopes.
Plos Biol., 19, 2021
|
|
8TNM
 
 | |
8UG1
 
 | | Crystal structure of KHK-C and compound 13 | | Descriptor: | GLYCEROL, Ketohexokinase, SULFATE ION, ... | | Authors: | Durbin, J.D, Guo, S.Y. | | Deposit date: | 2023-10-05 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Identification of LY3522348: A Highly Selective and Orally Efficacious Ketohexokinase Inhibitor.
J.Med.Chem., 66, 2023
|
|
8UG3
 
 | | Crystal structure of KHK-C and compound 23 | | Descriptor: | 2-[(4P)-4-{2-[(2S)-2-methylazetidin-1-yl]-6-(trifluoromethyl)pyrimidin-4-yl}-1H-pyrazol-1-yl]-1-(piperazin-1-yl)ethan-1-one, GLYCEROL, Ketohexokinase, ... | | Authors: | Durbin, J.D, Guo, S.Y. | | Deposit date: | 2023-10-05 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Identification of LY3522348: A Highly Selective and Orally Efficacious Ketohexokinase Inhibitor.
J.Med.Chem., 66, 2023
|
|
4LOH
 
 | | Crystal structure of hSTING(H232) in complex with c[G(2',5')pA(3',5')p] | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Function Analysis of STING Activation by c[G(2',5')pA(3',5')p] and Targeting by Antiviral DMXAA.
Cell(Cambridge,Mass.), 154, 2013
|
|
4LOJ
 
 | | Crystal structure of mSting in complex with c[G(2',5')pA(3',5')p] | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Gao, P, Patel, D.J. | | Deposit date: | 2013-07-12 | | Release date: | 2013-08-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure-Function Analysis of STING Activation by c[G(2',5')pA(3',5')p] and Targeting by Antiviral DMXAA.
Cell(Cambridge,Mass.), 154, 2013
|
|
3UAT
 
 | |
3FEP
 
 | | Crystal structure of the R132K:R111L:L121E:R59W-CRABPII mutant complexed with a synthetic ligand (merocyanin) at 2.60 angstrom resolution. | | Descriptor: | (2E,4E,6E)-3-methyl-6-(1,3,3-trimethyl-1,3-dihydro-2H-indol-2-ylidene)hexa-2,4-dienal, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cellular retinoic acid-binding protein 2 | | Authors: | Jia, X, Geiger, J.H. | | Deposit date: | 2008-11-30 | | Release date: | 2009-11-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | "Turn-on" protein fluorescence: in situ formation of cyanine dyes.
J.Am.Chem.Soc., 137, 2015
|
|
3FPM
 
 | | Crystal Structure of a Squarate Inhibitor bound to MAPKAP Kinase-2 | | Descriptor: | 3-{[(1R)-1-phenylethyl]amino}-4-(pyridin-4-ylamino)cyclobut-3-ene-1,2-dione, MAP kinase-activated protein kinase 2 | | Authors: | Parris, K.D. | | Deposit date: | 2009-01-05 | | Release date: | 2009-04-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Identification and SAR of squarate inhibitors of mitogen activated protein kinase-activated protein kinase 2 (MK-2)
Bioorg.Med.Chem., 17, 2009
|
|
7T1F
 
 | | Crystal structure of GDP-bound T50I mutant of human KRAS4B | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, MAGNESIUM ION | | Authors: | Zhang, Y, Zhang, C. | | Deposit date: | 2021-12-01 | | Release date: | 2022-12-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and functional analyses of a germline KRAS T50I mutation provide insights into Raf activation.
JCI Insight, 8, 2023
|
|
