7MZ9
 
 | | Cryo-EM structure of minimal TRPV1 with 1 partially bound RTX | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7MZB
 
 | | Cryo-EM structure of minimal TRPV1 with 3 bound RTX and 1 perturbed PI | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7MZC
 
 | |
7MZA
 
 | | Cryo-EM structure of minimal TRPV1 with 2 bound RTX in adjacent pockets | | Descriptor: | (2S)-1-(butanoyloxy)-3-{[(R)-hydroxy{[(1r,2R,3S,4S,5R,6S)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}propan-2-yl tridecanoate, SODIUM ION, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Zhang, K, Julius, D, Cheng, Y. | | Deposit date: | 2021-05-24 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Structural snapshots of TRPV1 reveal mechanism of polymodal functionality.
Cell, 184, 2021
|
|
7MZ7
 
 | |
7MZD
 
 | |
7XN5
 
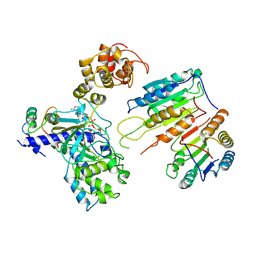 | | Cryo-EM structure of CopC-CaM-caspase-3 with ADPR | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Arginine ADP-riboxanase CopC, Calmodulin-1, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
8CXO
 
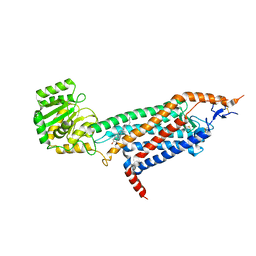 | | Cryo-EM structure of the unliganded mSMO-PGS2 in a lipidic environment | | Descriptor: | CHOLESTEROL, Smoothened homolog, GlgA glycogen synthase chimera | | Authors: | Zhang, K, Wu, H, Hoppe, N, Manglik, A, Cheng, Y. | | Deposit date: | 2022-05-22 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Fusion protein strategies for cryo-EM study of G protein-coupled receptors.
Nat Commun, 13, 2022
|
|
7X01
 
 | | Cryo-EM Structure of Chikungunya Virus Nonstructural Protein 1 with inhibitor FHA | | Descriptor: | (1R,2S,3S,4R,5R)-3-(6-aminopurin-9-yl)-4-fluoranyl-5-(2-hydroxyethyl)cyclopentane-1,2-diol, ZINC ION, mRNA-capping enzyme nsP1 | | Authors: | Zhang, K, Law, M.C.Y, Nguyen, T.M, Tan, Y.B, Wirawan, M, Law, Y.S, Luo, D.H. | | Deposit date: | 2022-02-20 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Molecular basis of specific viral RNA recognition and 5'-end capping by the Chikungunya virus nsP1.
Cell Rep, 40, 2022
|
|
7XN6
 
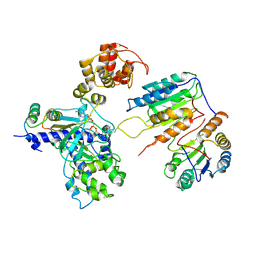 | | Cryo-EM structure of CopC-CaM-caspase-3 with ADPR-deacylization | | Descriptor: | Arginine ADP-riboxanase CopC, Calmodulin-1, Caspase-3, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
7XN4
 
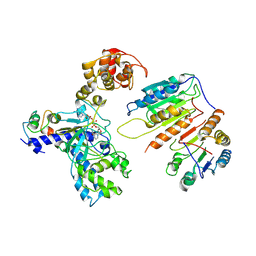 | | Cryo-EM structure of CopC-CaM-caspase-3 with NAD+ | | Descriptor: | Arginine ADP-riboxanase CopC, Calmodulin-1, Caspase-3, ... | | Authors: | Zhang, K, Peng, T, Tao, X.Y, Tian, M, Li, Y.X, Wang, Z, Ma, S.F, Hu, S.F, Pan, X, Xue, J, Luo, J.W, Wu, Q.L, Fu, Y, Li, S. | | Deposit date: | 2022-04-28 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structural insights into caspase ADPR deacylization catalyzed by a bacterial effector and host calmodulin.
Mol.Cell, 82, 2022
|
|
7DOP
 
 | | Structural insights into viral RNA capping and plasma membrane targeting by Chikungunya virus nonstructural protein 1 | | Descriptor: | Nonstructural Protein 1, ZINC ION | | Authors: | Zhang, K, Law, Y.S, Law, M.C.Y, Tan, Y.B, Wirawan, M, Luo, D.H. | | Deposit date: | 2020-12-15 | | Release date: | 2021-03-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.38 Å) | | Cite: | Structural insights into viral RNA capping and plasma membrane targeting by Chikungunya virus nonstructural protein 1.
Cell Host Microbe, 29, 2021
|
|
7FGG
 
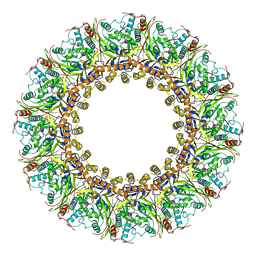 | | Cryo-EM Structure of Chikungunya Virus Nonstructural Protein 1 with m7GTP | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Zhang, K, Law, M.C.Y, Nguyen, T.M, Tan, Y.B, Wirawan, M, Law, Y.S, Luo, D.H. | | Deposit date: | 2021-07-27 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Molecular basis of specific viral RNA recognition and 5'-end capping by the Chikungunya virus nsP1.
Cell Rep, 40, 2022
|
|
7FGH
 
 | | Cryo-EM Structure of Chikungunya Virus Nonstructural Protein 1 with m7GMP | | Descriptor: | N7-METHYL-GUANOSINE-5'-MONOPHOSPHATE, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Zhang, K, Law, M.C.Y, Nguyen, T.M, Tan, Y.B, Wirawan, M, Law, Y.S, Luo, D.H. | | Deposit date: | 2021-07-27 | | Release date: | 2022-07-27 | | Last modified: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (2.18 Å) | | Cite: | Molecular basis of specific viral RNA recognition and 5'-end capping by the Chikungunya virus nsP1.
Cell Rep, 40, 2022
|
|
7FGI
 
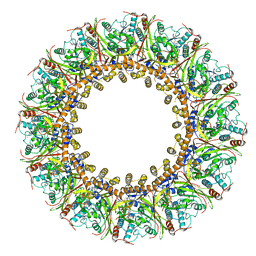 | | Cryo-EM Structure of Chikungunya Virus Nonstructural Protein 1 with m7Gppp-AU | | Descriptor: | MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION, ... | | Authors: | Zhang, K, Law, M.C.Y, Nguyen, T.M, Tan, Y.B, Wirawan, M, Law, Y.S, Luo, D.H. | | Deposit date: | 2021-07-27 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Molecular basis of specific viral RNA recognition and 5'-end capping by the Chikungunya virus nsP1.
Cell Rep, 40, 2022
|
|
7BZ0
 
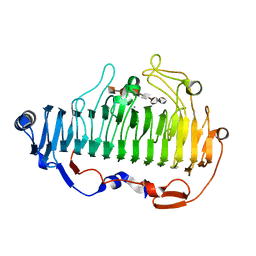 | | complex structure of alginate lyase AlyF-OU02 with G6 | | Descriptor: | Alginate lyase AlyF-OU02, CALCIUM ION, alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid-(1-4)-alpha-L-gulopyranuronic acid | | Authors: | Liu, W, Lyu, Q, Zhang, K. | | Deposit date: | 2020-04-26 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the substrate-binding cleft of AlyF reveal the first long-chain alginate-binding mode.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
5ADX
 
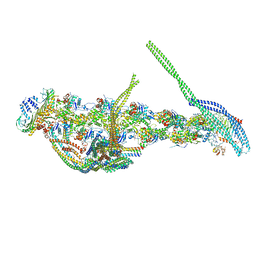 | | CryoEM structure of dynactin complex at 4.0 angstrom resolution | | Descriptor: | ACTIN RELATED PROTEIN 1, ACTIN RELATED PROTEIN 11, ACTIN, ... | | Authors: | Zhang, K, Urnavicius, L, Diamant, A.G, Motz, C, Schlage, M.A, Yu, M, Patel, N.A, Robinson, C.V, Carter, A.P. | | Deposit date: | 2015-08-24 | | Release date: | 2015-12-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The Structure of the Dynactin Complex and its Interaction with Dynein.
Science, 347, 2015
|
|
6D00
 
 | | Calcarisporiella thermophila Hsp104 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Calcarisporiella thermophila Hsp104 | | Authors: | Zhang, K, Pintilie, G. | | Deposit date: | 2018-04-09 | | Release date: | 2019-04-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of Calcarisporiella thermophila Hsp104 Disaggregase that Antagonizes Diverse Proteotoxic Misfolding Events.
Structure, 27, 2019
|
|
8QQ3
 
 | | Streptavidin with a Ni-cofactor | | Descriptor: | 4-[4-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]butylamino]-~{N}1,~{N}1'-di(quinolin-8-yl)cyclohexane-1,1-dicarboxamide, NICKEL (II) ION, Streptavidin | | Authors: | Zhang, K, Jakob, R.P, Ward, T.R. | | Deposit date: | 2023-10-03 | | Release date: | 2024-02-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An artificial nickel chlorinase based on the biotin-streptavidin technology.
Chem.Commun.(Camb.), 60, 2024
|
|
6NUD
 
 | | Small conformation of ssRNA-bound CRISPR_Csm complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cms protein Csm2, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), ... | | Authors: | Zhang, K, Pintilie, G, Li, S, Zhu, Y, Chiu, W, Huang, Z. | | Deposit date: | 2019-01-31 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Coupling of ssRNA cleavage with DNase activity in type III-A CRISPR-Csm revealed by cryo-EM and biochemistry.
Cell Res., 29, 2019
|
|
6NUE
 
 | | Small conformation of apo CRISPR_Csm complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CRISPR system Cms protein Csm2, CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A), ... | | Authors: | Zhang, K, Pintilie, G, Li, S, Zhu, Y, Chiu, W, Huang, Z. | | Deposit date: | 2019-01-31 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Coupling of ssRNA cleavage with DNase activity in type III-A CRISPR-Csm revealed by cryo-EM and biochemistry.
Cell Res., 29, 2019
|
|
6WQH
 
 | | Molecular basis for the ATPase-powered substrate translocation by the Lon AAA+ protease | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Ig2 substrate, Lon protease, ... | | Authors: | Zhang, K, Li, S, Hsiehb, K, Sub, S, Pintilie, G, Chiu, W, Chang, C. | | Deposit date: | 2020-04-28 | | Release date: | 2021-06-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Molecular basis for ATPase-powered substrate translocation by the Lon AAA+ protease.
J.Biol.Chem., 297, 2021
|
|
7RRP
 
 | |
3EPU
 
 | | Crystal Structure of STM2138, a novel virulence chaperone in Salmonella | | Descriptor: | STM2138 Virulence Chaperone | | Authors: | Zhang, K, Andres, S.N, Hannemann, M, Coombes, B, Junop, M. | | Deposit date: | 2008-09-30 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural analysis and quantitative proteomic
interactome of a novel virulence chaperone in Salmonella
To be Published
|
|
2GMW
 
 | | Crystal Structure of D,D-heptose 1.7-bisphosphate phosphatase from E. Coli. | | Descriptor: | D,D-heptose 1,7-bisphosphate phosphatase, ZINC ION | | Authors: | Zhang, K, DeLeon, G, Wright, G.D, Junop, M.S. | | Deposit date: | 2006-04-07 | | Release date: | 2007-04-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and kinetic characterization of the LPS biosynthetic enzyme D-alpha,beta-D-heptose-1,7-bisphosphate phosphatase (GmhB) from Escherichia coli.
Biochemistry, 49, 2010
|
|
