4WNW
 
 | | Human Cytochrome P450 2D6 Thioridazine Complex | | Descriptor: | 10-{2-[(2R)-1-methylpiperidin-2-yl]ethyl}-2-(methylsulfanyl)-10H-phenothiazine, Cytochrome P450 2D6, NICKEL (II) ION, ... | | Authors: | Wang, A, Stout, C.D, Johnson, E.F. | | Deposit date: | 2014-10-14 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.299 Å) | | Cite: | Contributions of Ionic Interactions and Protein Dynamics to Cytochrome P450 2D6 (CYP2D6) Substrate and Inhibitor Binding.
J.Biol.Chem., 290, 2015
|
|
4WNV
 
 | | Human Cytochrome P450 2D6 Quinine Complex | | Descriptor: | Cytochrome P450 2D6, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wang, A, Stout, C.D, Johnson, E.F. | | Deposit date: | 2014-10-14 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Contributions of Ionic Interactions and Protein Dynamics to Cytochrome P450 2D6 (CYP2D6) Substrate and Inhibitor Binding.
J.Biol.Chem., 290, 2015
|
|
4WNU
 
 | | Human Cytochrome P450 2D6 Quinidine Complex | | Descriptor: | Cytochrome P450 2D6, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Wang, A, Stout, C.D, Johnson, E.F. | | Deposit date: | 2014-10-14 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Contributions of Ionic Interactions and Protein Dynamics to Cytochrome P450 2D6 (CYP2D6) Substrate and Inhibitor Binding.
J.Biol.Chem., 290, 2015
|
|
6FDR
 
 | | 7-FE FERREDOXIN FROM AZOTOBACTER VINELANDII AT 100K, NA DITHIONITE REDUCED AT PH 8.5, RESOLUTION 1.4 A | | Descriptor: | FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, PROTEIN (7-FE FERREDOXIN I) | | Authors: | Schipke, C.G, Goodin, D.B, Mcree, D.E, Stout, C.D. | | Deposit date: | 1998-12-11 | | Release date: | 1998-12-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Oxidized and reduced Azotobacter vinelandii ferredoxin I at 1.4 A resolution: conformational change of surface residues without significant change in the [3Fe-4S]+/0 cluster.
Biochemistry, 38, 1999
|
|
4WNT
 
 | | Human Cytochrome P450 2D6 Ajmalicine Complex | | Descriptor: | Ajmalicine, Cytochrome P450 2D6, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Wang, A, Stout, C.D, Johnson, E.F. | | Deposit date: | 2014-10-14 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Contributions of Ionic Interactions and Protein Dynamics to Cytochrome P450 2D6 (CYP2D6) Substrate and Inhibitor Binding.
J.Biol.Chem., 290, 2015
|
|
2BDM
 
 | | Structure of Cytochrome P450 2B4 with Bound Bifonazole | | Descriptor: | 1-[PHENYL-(4-PHENYLPHENYL)-METHYL]IMIDAZOLE, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, ... | | Authors: | Zhao, Y, White, M.A, Muralidhara, B.K, Sun, L, Halpert, J.R, Stout, C.D. | | Deposit date: | 2005-10-20 | | Release date: | 2005-12-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of microsomal cytochrome P450 2B4 complexed with the antifungal drug bifonazole: insight into P450 conformational plasticity and membrane interaction.
J.Biol.Chem., 281, 2006
|
|
1D3W
 
 | | Crystal structure of ferredoxin 1 d15e mutant from azotobacter vinelandii at 1.7 angstrom resolution. | | Descriptor: | FE3-S4 CLUSTER, FERREDOXIN 1, IRON/SULFUR CLUSTER | | Authors: | Chen, K, Hirst, J, Camba, R, Bonagura, C.A, Stout, C.D, Burges, B.K, Armstrong, F.A. | | Deposit date: | 1999-10-01 | | Release date: | 1999-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Atomically defined mechanism for proton transfer to a buried redox centre in a protein.
Nature, 405, 2000
|
|
1LBE
 
 | | APLYSIA ADP RIBOSYL CYCLASE | | Descriptor: | ADP RIBOSYL CYCLASE | | Authors: | Prasad, G.S, Mcree, D.E, Stura, E.A, Levitt, D.G, Lee, H.C, Stout, C.D. | | Deposit date: | 1996-09-18 | | Release date: | 1997-09-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Aplysia ADP ribosyl cyclase, a homologue of the bifunctional ectozyme CD38.
Nat.Struct.Biol., 3, 1996
|
|
1LYN
 
 | |
5W5W
 
 | |
5VJ3
 
 | |
2VN0
 
 | | CYP2C8DH COMPLEXED WITH TROGLITAZONE | | Descriptor: | (5R)-5-(4-{[(2R)-6-HYDROXY-2,5,7,8-TETRAMETHYL-3,4-DIHYDRO-2H-CHROMEN-2-YL]METHOXY}BENZYL)-1,3-THIAZOLIDINE-2,4-DIONE, CYTOCHROME P450 2C8, PALMITIC ACID, ... | | Authors: | Schoch, G.A, Yano, J.K, Sansen, S, Stout, C.D, Johnson, E.F. | | Deposit date: | 2008-01-30 | | Release date: | 2008-04-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Determinants of cytochrome P450 2C8 substrate binding: structures of complexes with montelukast, troglitazone, felodipine, and 9-cis-retinoic acid.
J. Biol. Chem., 283, 2008
|
|
3MDM
 
 | | Thioperamide complex of Cytochrome P450 46A1 | | Descriptor: | Cholesterol 24-hydroxylase, N-cyclohexyl-4-(1H-imidazol-5-yl)piperidine-1-carbothioamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mast, N, Charvet, C, Pikuleva, I, Stout, C.D. | | Deposit date: | 2010-03-30 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of drug binding to CYP46A1, an enzyme that controls cholesterol turnover in the brain.
J.Biol.Chem., 285, 2010
|
|
3MDV
 
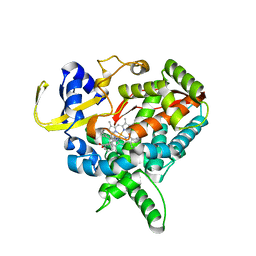 | | Clotrimazole complex of Cytochrome P450 46A1 | | Descriptor: | 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, Cholesterol 24-hydroxylase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mast, N, Charvet, C, Pikuleva, I, Stout, C.D. | | Deposit date: | 2010-03-30 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of drug binding to CYP46A1, an enzyme that controls cholesterol turnover in the brain.
J.Biol.Chem., 285, 2010
|
|
2Q9G
 
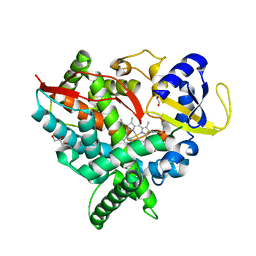 | | Crystal structure of human cytochrome P450 46A1 | | Descriptor: | Cytochrome P450 46A1, GLYCEROL, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | White, M.A, Mast, N.V, Johnson, E.F, Stout, C.D, Pikuleva, I.A. | | Deposit date: | 2007-06-12 | | Release date: | 2008-06-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of substrate-bound and substrate-free cytochrome P450 46A1, the principal cholesterol hydroxylase in the brain.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2Q9F
 
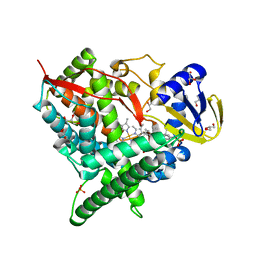 | | Crystal structure of human cytochrome P450 46A1 in complex with cholesterol-3-sulphate | | Descriptor: | CHOLEST-5-EN-3-YL HYDROGEN SULFATE, Cytochrome P450 46A1, GLYCEROL, ... | | Authors: | White, M.A, Mast, N.V, Johnson, E.F, Stout, C.D, Pikuleva, I.A. | | Deposit date: | 2007-06-12 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of substrate-bound and substrate-free cytochrome P450 46A1, the principal cholesterol hydroxylase in the brain.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1C97
 
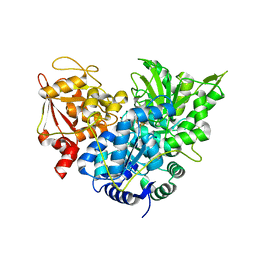 | | S642A:ISOCITRATE COMPLEX OF ACONITASE | | Descriptor: | IRON/SULFUR CLUSTER, ISOCITRIC ACID, MITOCHONDRIAL ACONITASE, ... | | Authors: | Lloyd, S.J, Lauble, H, Prasad, G.S, Stout, C.D. | | Deposit date: | 1999-07-31 | | Release date: | 1999-08-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | The mechanism of aconitase: 1.8 A resolution crystal structure of the S642a:citrate complex.
Protein Sci., 8, 1999
|
|
1D4O
 
 | | CRYSTAL STRUCTURE OF TRANSHYDROGENASE DOMAIN III AT 1.2 ANGSTROMS RESOLUTION | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP(H) TRANSHYDROGENASE | | Authors: | Prasad, G.S, Sridhar, V, Yamaguchi, M, Hatefi, Y, Stout, C.D. | | Deposit date: | 1999-10-04 | | Release date: | 2000-01-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Crystal structure of transhydrogenase domain III at 1.2 A resolution.
Nat.Struct.Biol., 6, 1999
|
|
1DUT
 
 | | FIV DUTP PYROPHOSPHATASE | | Descriptor: | DUTP PYROPHOSPHATASE, MAGNESIUM ION | | Authors: | Prasad, G.S, Stura, E.A, Mcree, D.E, Laco, G.S, Hasselkus-Light, C, Elder, J.H, Stout, C.D. | | Deposit date: | 1996-09-15 | | Release date: | 1997-01-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of dUTP pyrophosphatase from feline immunodeficiency virus.
Protein Sci., 5, 1996
|
|
4N4Y
 
 | |
4O9U
 
 | | Mechanism of transhydrogenase coupling proton translocation and hydride transfer | | Descriptor: | NAD(P) transhydrogenase subunit alpha 2, NAD(P) transhydrogenase subunit beta, NAD/NADP transhydrogenase alpha subunit 1, ... | | Authors: | Leung, J.H, Yamaguchi, M, Moeller, A, Schurig-Briccio, L.A, Gennis, R.B, Potter, C.S, Carragher, B, Stout, C.D. | | Deposit date: | 2014-01-02 | | Release date: | 2015-01-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (6.926 Å) | | Cite: | Structural biology. Division of labor in transhydrogenase by alternating proton translocation and hydride transfer.
Science, 347, 2015
|
|
4O9P
 
 | | Crystal structure of Thermus thermophilis transhydrogeanse domain II dimer SeMet derivative | | Descriptor: | NAD(P) transhydrogenase subunit alpha 2, NAD(P) transhydrogenase subunit beta | | Authors: | Leung, J.H, Yamaguchi, M, Moeller, A, Schurig-Briccio, L.A, Gennis, R.B, Potter, C.S, Carragher, B, Stout, C.D. | | Deposit date: | 2014-01-02 | | Release date: | 2014-06-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural biology. Division of labor in transhydrogenase by alternating proton translocation and hydride transfer.
Science, 347, 2015
|
|
5E0E
 
 | |
5EM4
 
 | | Structure of CYP2B4 F244W in a ligand free conformation | | Descriptor: | 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Shah, M.B, Stout, C.D, Halpert, J.R. | | Deposit date: | 2015-11-05 | | Release date: | 2016-03-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Coumarin Derivatives as Substrate Probes of Mammalian Cytochromes P450 2B4 and 2B6: Assessing the Importance of 7-Alkoxy Chain Length, Halogen Substitution, and Non-Active Site Mutations.
Biochemistry, 55, 2016
|
|
5VCK
 
 | | HIV Protease (PR) with TL-3 in the active site and (Z)-N-(thiazol-2-yl)-N'-tosylcarbamimidate in the exosite | | Descriptor: | 4-methyl-N-(thiazol-2-ylcarbamoyl)benzenesulfonamide, Protease, benzyl [(1S,4S,7S,8R,9R,10S,13S,16S)-7,10-dibenzyl-8,9-dihydroxy-1,16-dimethyl-4,13-bis(1-methylethyl)-2,5,12,15,18-pentaoxo-20-phenyl-19-oxa-3,6,11,14,17-pentaazaicos-1-yl]carbamate | | Authors: | Tiefenbrunn, T, Stout, C.D. | | Deposit date: | 2017-03-31 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fragment-based campaign for the identification of potential exosite binders of HIV-1 Protease
to be published
|
|
