1W6K
 
 | | Structure of human OSC in complex with Lanosterol | | Descriptor: | LANOSTEROL, LANOSTEROL SYNTHASE, octyl beta-D-glucopyranoside | | Authors: | Thoma, R, Schulz-Gasch, T, D'Arcy, B, Benz, J, Aebi, J, Dehmlow, H, Hennig, M, Ruf, A. | | Deposit date: | 2004-08-19 | | Release date: | 2004-10-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Insight Into Steroid Scaffold Formation from the Structure of Human Oxidosqualene Cyclase
Nature, 432, 2004
|
|
1W6J
 
 | | Structure of human OSC in complex with Ro 48-8071 | | Descriptor: | LANOSTEROL SYNTHASE, TETRADECANE, [4-({6-[ALLYL(METHYL)AMINO]HEXYL}OXY)-2-FLUOROPHENYL](4-BROMOPHENYL)METHANONE, ... | | Authors: | Thoma, R, Schulz-Gasch, T, D'Arcy, B, Benz, J, Aebi, J, Dehmlow, H, Hennig, M, Ruf, A. | | Deposit date: | 2004-08-18 | | Release date: | 2004-10-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insight Into Steroid Scaffold Formation from the Structure of Human Oxidosqualene Cyclase
Nature, 432, 2004
|
|
2FVJ
 
 | | A novel anti-adipogenic partial agonist of peroxisome proliferator-activated receptor-gamma (PPARG) recruits pparg-coactivator-1 alpha (PGC1A) but potentiates insulin signaling in vitro | | Descriptor: | 1-(3,4-DIMETHOXYBENZYL)-6,7-DIMETHOXY-4-{[4-(2-METHOXYPHENYL)PIPERIDIN-1-YL]METHYL}ISOQUINOLINE, GLYCEROL, Nuclear receptor coactivator 1, ... | | Authors: | Benz, J, Burgermeister, E, Flament, A, Schnoebelen, A, Stihle, M, Gsell, B, Rufer, A, Ruf, A, Kuhn, B, Maerki, H.P, Mizrahi, J, Sebokova, E, Niesor, E, Meyer, M. | | Deposit date: | 2006-01-31 | | Release date: | 2006-05-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A novel partial agonist of peroxisome proliferator-activated receptor-gamma (PPARgamma) recruits PPARgamma-coactivator-1alpha, prevents triglyceride accumulation, and potentiates insulin signaling in vitro
Mol.Endocrinol., 20, 2006
|
|
3SGJ
 
 | | Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgIII and antibodies lacking core fucose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MALONATE ION, ... | | Authors: | Ferrara, C, Grau, S, Jaeger, C, Sondermann, P, Bruenker, P, Waldhauer, I, Hennig, M, Ruf, A, Rufer, A.C, Stihle, M, Umana, P, Benz, J. | | Deposit date: | 2011-06-15 | | Release date: | 2011-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgammaRIII and antibodies lacking core fucose.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3SGK
 
 | | Unique carbohydrate/carbohydrate interactions are required for high affinity binding of FcgIII and antibodies lacking core fucose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)][2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fc fragment, ... | | Authors: | Ferrara, C, Grau, S, Jaeger, C, Sondermann, P, Bruenker, P, Waldhauer, I, Hennig, M, Ruf, A, Rufer, A.C, Stihle, M, Umana, P, Benz, J. | | Deposit date: | 2011-06-15 | | Release date: | 2011-08-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgammaRIII and antibodies lacking core fucose.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3MNP
 
 | | Crystal structure of the agonist form of mouse glucocorticoid receptor stabilized by (A611V, V708A, E711G) mutations at 1.50A | | Descriptor: | DEXAMETHASONE, GLYCEROL, Glucocorticoid receptor, ... | | Authors: | Schoch, G.A, Seitz, T, Benz, J, Banner, D, Stihle, M, D'Arcy, B, Thoma, R, Sterner, R, Hennig, M, Ruf, A. | | Deposit date: | 2010-04-22 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Enhancing the stability and solubility of the glucocorticoid receptor ligand-binding domain by high-throughput library screening.
J.Mol.Biol., 403, 2010
|
|
3MNE
 
 | | Crystal structure of the agonist form of mouse glucocorticoid receptor stabilized by F608S mutation at 1.96A | | Descriptor: | DEXAMETHASONE, GLYCEROL, Glucocorticoid receptor, ... | | Authors: | Schoch, G.A, Seitz, T, Benz, J, Banner, D, Stihle, M, D'Arcy, B, Thoma, R, Sterner, R, Hennig, M, Ruf, A. | | Deposit date: | 2010-04-21 | | Release date: | 2010-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Enhancing the stability and solubility of the glucocorticoid receptor ligand-binding domain by high-throughput library screening.
J.Mol.Biol., 403, 2010
|
|
3ZKX
 
 | | TERNARY BACE2 XAPERONE COMPLEX | | Descriptor: | BETA-SECRETASE 2, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Kuglstatter, A, Banner, D.W, Benz, J, Bertschinger, J, Burger, D, Cuppuleri, S, Debulpaep, M, Gast, A, Grabulovski, D, Gsell, B, Hilpert, H, Huber, W, Kusznir, E, Laeremans, T, Matile, H, Rufer, A, Schlatter, D, Steyeart, J, Stihle, M, Thoma, R, Weber, M, Ruf, A. | | Deposit date: | 2013-01-25 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3G8I
 
 | | Aleglitazar, a new, potent, and balanced PPAR alpha/gamma agonist for the treatment of type II diabetes | | Descriptor: | (2S)-2-methoxy-3-{4-[2-(5-methyl-2-phenyl-1,3-oxazol-4-yl)ethoxy]-1-benzothiophen-7-yl}propanoic acid, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor alpha | | Authors: | Benz, J, Bernardeau, A, Binggeli, A, Blum, D, Boehringer, M, Grether, U, Hilpert, H, Kuhn, B, Maerki, H.P, Meyer, M, Puentener, K, Raab, S, Ruf, A, Schlatter, D, Gsell, B, Stihle, M, Mohr, P. | | Deposit date: | 2009-02-12 | | Release date: | 2009-06-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Aleglitazar, a new, potent, and balanced dual PPARalpha/gamma agonist for the treatment of type II diabetes.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3FEI
 
 | | Design and biological evaluation of novel, balanced dual PPARa/g agonists | | Descriptor: | (2S)-3-(4-{[2-(4-chlorophenyl)-1,3-thiazol-4-yl]methoxy}-2-methylphenyl)-2-ethoxypropanoic acid, Peptide motif 5 of Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor alpha | | Authors: | Benz, J, Grether, U, Gsell, B, Binggeli, A, Hilpert, H, Maerki, H.P, Mohr, P, Ruf, A, Stihle, M, Schlatter, D. | | Deposit date: | 2008-11-30 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and biological evaluation of novel, balanced dual PPARalpha/gamma agonists
Chemmedchem, 4, 2009
|
|
3FEJ
 
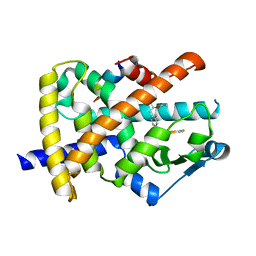 | | Design and biological evaluation of novel, balanced dual PPARa/g agonists | | Descriptor: | (2S)-3-(4-{[2-(4-chlorophenyl)-1,3-thiazol-4-yl]methoxy}-2-methylphenyl)-2-ethoxypropanoic acid, Peroxisome proliferator-activated receptor gamma, peptide motif 3 of Nuclear receptor coactivator 1 | | Authors: | Benz, J, Grether, U, Gsell, B, Binggeli, A, Hilpert, H, Kuhn, B, Maerki, H.P, Mohr, P, Ruf, A, Stihle, M. | | Deposit date: | 2008-11-30 | | Release date: | 2009-10-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Design and biological evaluation of novel, balanced dual PPARalpha/gamma agonists
Chemmedchem, 4, 2009
|
|
1NU8
 
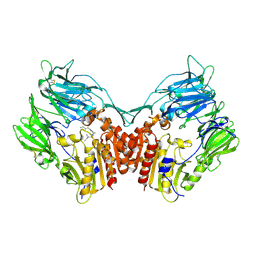 | | Crystal structure of human dipeptidyl peptidase IV (DPP-IV) in complex with Diprotin A (IPI) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-mer peptide, Dipeptidyl peptidase IV | | Authors: | Thoma, R, Loeffler, B, Stihle, M, Huber, W, Ruf, A, Hennig, M. | | Deposit date: | 2003-01-31 | | Release date: | 2003-08-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of Proline-Specific Exopeptidase Activity as Observed in Human Dipeptidyl Peptidase-IV.
Structure, 11, 2003
|
|
3ZKM
 
 | | BACE2 FAB COMPLEX | | Descriptor: | BETA-SECRETASE 2, DIMETHYL SULFOXIDE, FAB HEAVY CHAIN, ... | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-01-23 | | Release date: | 2013-05-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3ZOV
 
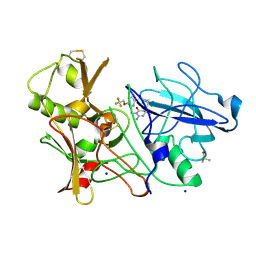 | | CRYSTAL STRUCTURE OF BACE-1 IN COMPLEX WITH CHEMICAL LIGAND | | Descriptor: | 5-(2,2,2-Trifluoro-ethoxy)-pyridine-2-carboxylic acid [3-((S)-2-amino-1,4-dimethyl-6-oxo-1,4,5,6-tetrahydro-pyrimidin-4-yl)-phenyl]-amide, BETA-SECRETASE 1, DIMETHYL SULFOXIDE, ... | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-02-25 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3ZKQ
 
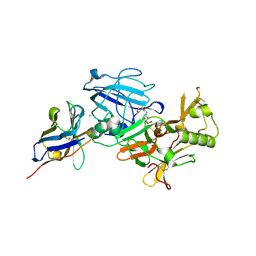 | | BACE2 XAPERONE COMPLEX | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-SECRETASE 2, CHLORIDE ION, ... | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-01-24 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3ZKI
 
 | | BACE2 MUTANT STRUCTURE WITH LIGAND | | Descriptor: | 5-(2,2,2-Trifluoro-ethoxy)-pyridine-2-carboxylic acid [3-((S)-2-amino-1,4-dimethyl-6-oxo-1,4,5,6-tetrahydro-pyrimidin-4-yl)-phenyl]-amide, BETA-SECRETASE 2 | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-01-23 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3ZKG
 
 | | BACE2 MUTANT APO STRUCTURE | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-SECRETASE 2 | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-01-23 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3ZL7
 
 | | BACE2 FYNOMER COMPLEX | | Descriptor: | BETA-SECRETASE 2, FYNOMER 2B-H11 | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-01-28 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
1NU6
 
 | | Crystal structure of human Dipeptidyl Peptidase IV (DPP-IV) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase IV, MERCURY (II) ION | | Authors: | Hennig, M, Stihle, M, Thoma, R, Ruf, A. | | Deposit date: | 2003-01-31 | | Release date: | 2003-08-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Basis of Proline-Specific Exopeptidase Activity as Observed in Human Dipeptidyl Peptidase-IV.
Structure, 11, 2003
|
|
3ZKS
 
 | | BACE2 XAPERONE COMPLEX WITH INHIBITOR | | Descriptor: | 5-(2,2,2-Trifluoro-ethoxy)-pyridine-2-carboxylic acid [3-((S)-2-amino-1,4-dimethyl-6-oxo-1,4,5,6-tetrahydro-pyrimidin-4-yl)-phenyl]-amide, BETA-SECRETASE 2, XA4813 | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-01-24 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3ZKN
 
 | | BACE2 FAB INHIBITOR COMPLEX | | Descriptor: | 5-(2,2,2-Trifluoro-ethoxy)-pyridine-2-carboxylic acid [3-((S)-2-amino-1,4-dimethyl-6-oxo-1,4,5,6-tetrahydro-pyrimidin-4-yl)-phenyl]-amide, BETA-SECRETASE 2, DIMETHYL SULFOXIDE, ... | | Authors: | Banner, D.W, Kuglstatter, A, Benz, J, Stihle, M, Ruf, A. | | Deposit date: | 2013-01-23 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Mapping the Conformational Space Accessible to Bace2 Using Surface Mutants and Co-Crystals with Fab-Fragments, Fynomers, and Xaperones
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3S3X
 
 | | Structure of chicken acid-sensing ion channel 1 AT 3.0 A resolution in complex with psalmotoxin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Amiloride-sensitive cation channel 2, neuronal, ... | | Authors: | Dawson, R.J.P, Benz, J, Stohler, P, Tetaz, T, Joseph, C, Huber, S, Schmid, G, Huegin, D, Pflimlin, P, Trube, G, Rudolph, M.G, Hennig, M, Ruf, A. | | Deposit date: | 2011-05-18 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structure of the Acid-sensing ion channel 1 in complex with the gating modifier Psalmotoxin 1.
Nat Commun, 3, 2012
|
|
3S3W
 
 | | Structure of chicken acid-sensing ion channel 1 at 2.6 a resolution and ph 7.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Amiloride-sensitive cation channel 2, ... | | Authors: | Dawson, R.J.P, Benz, J, Stohler, P, Tetaz, T, Joseph, C, Huber, S, Schmid, G, Huegin, D, Pflimlin, P, Trube, G, Rudolph, M.G, Hennig, M, Ruf, A. | | Deposit date: | 2011-05-18 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of the Acid-sensing ion channel 1 in complex with the gating modifier Psalmotoxin 1.
Nat Commun, 3, 2012
|
|
3H52
 
 | | Crystal structure of the antagonist form of human glucocorticoid receptor | | Descriptor: | 11-(4-DIMETHYLAMINO-PHENYL)-17-HYDROXY-13-METHYL-17-PROP-1-YNYL-1,2,6,7,8,11,12,13,14,15,16,17-DODEC AHYDRO-CYCLOPENTA[A]PHENANTHREN-3-ONE, GLYCEROL, Glucocorticoid receptor, ... | | Authors: | Schoch, G.A, Benz, J, D'Arcy, B, Stihle, M, Burger, D, Thoma, R, Ruf, A. | | Deposit date: | 2009-04-21 | | Release date: | 2009-12-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular switch in the glucocorticoid receptor: active and passive antagonist conformations
J.Mol.Biol., 395, 2010
|
|
4AFZ
 
 | | Human Chymase - Fynomer Complex | | Descriptor: | CHYMASE, D(-)-TARTARIC ACID, FYNOMER | | Authors: | Schlatter, D, Brack, S, Banner, D.W, Batey, S, Benz, J, Bertschinger, J, Huber, W, Joseph, C, Rufer, A, Van Der Kloosters, A, Weber, M, Grabulovski, D, Hennig, M. | | Deposit date: | 2012-01-23 | | Release date: | 2012-07-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Generation, Characterization and Structural Data of Chymase Binding Proteins Based on the Human Fyn Kinase SH3 Domain.
Mabs, 4, 2012
|
|
