3BVB
 
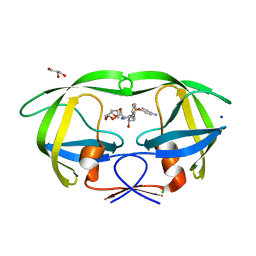 | | Cystal structure of HIV-1 Active Site Mutant D25N and inhibitor Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Liu, F, Weber, I.T. | | Deposit date: | 2008-01-05 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Effect of the Active Site D25N Mutation on the Structure, Stability, and Ligand Binding of the Mature HIV-1 Protease.
J.Biol.Chem., 283, 2008
|
|
3BVA
 
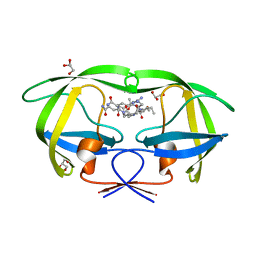 | | Cystal structure of HIV-1 Active Site Mutant D25N and p2-NC analog inhibitor | | Descriptor: | GLYCEROL, N-{(2S)-2-[(N-acetyl-L-threonyl-L-isoleucyl)amino]hexyl}-L-norleucyl-L-glutaminyl-N~5~-[amino(iminio)methyl]-L-ornithinamide, Protease (Retropepsin) | | Authors: | Liu, F, Weber, I.T. | | Deposit date: | 2008-01-05 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Effect of the Active Site D25N Mutation on the Structure, Stability, and Ligand Binding of the Mature HIV-1 Protease.
J.Biol.Chem., 283, 2008
|
|
4IGN
 
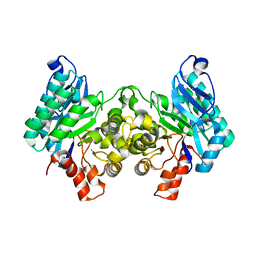 | | 2.32 Angstrom X-ray Crystal structure of R47A mutant of human ACMSD | | Descriptor: | 2-amino-3-carboxymuconate-6-semialdehyde decarboxylase, ZINC ION | | Authors: | Liu, F, Liu, A. | | Deposit date: | 2012-12-17 | | Release date: | 2014-05-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.329 Å) | | Cite: | Human alpha-amino-beta-carboxymuconate-epsilon-semialdehyde decarboxylase (ACMSD): A structural and mechanistic unveiling.
Proteins, 83, 2015
|
|
3KZ3
 
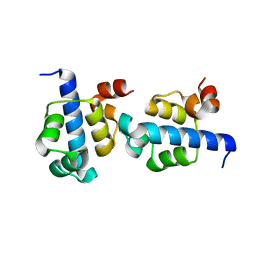 | |
3K5K
 
 | | Discovery of a 2,4-Diamino-7-aminoalkoxy-quinazoline as a Potent Inhibitor of Histone Lysine Methyltransferase, G9a | | Descriptor: | 7-[3-(dimethylamino)propoxy]-6-methoxy-2-(4-methyl-1,4-diazepan-1-yl)-N-(1-methylpiperidin-4-yl)quinazolin-4-amine, CHLORIDE ION, Histone-lysine N-methyltransferase, ... | | Authors: | Dong, A, Wasney, G.A, Liu, F, Chen, X, Allali-Hassani, A, Senisterra, G, Chau, I, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Frye, S.V, Bochkarev, A, Brown, P.J, Jin, J, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of a 2,4-diamino-7-aminoalkoxyquinazoline as a potent and selective inhibitor of histone lysine methyltransferase G9a.
J.Med.Chem., 52, 2009
|
|
2G69
 
 | |
1BOI
 
 | | N-TERMINALLY TRUNCATED RHODANESE | | Descriptor: | RHODANESE | | Authors: | Gliubich, F, Berni, R, Cianci, M, Trevino, R.J, Horowitz, P.M, Zanotti, G. | | Deposit date: | 1998-08-04 | | Release date: | 1999-04-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | NH2-terminal sequence truncation decreases the stability of bovine rhodanese, minimally perturbs its crystal structure, and enhances interaction with GroEL under native conditions.
J.Biol.Chem., 274, 1999
|
|
1BOH
 
 | | SULFUR-SUBSTITUTED RHODANESE (ORTHORHOMBIC FORM) | | Descriptor: | RHODANESE | | Authors: | Gliubich, F, Berni, R, Cianci, M, Trevino, R.J, Horowitz, P.M, Zanotti, G. | | Deposit date: | 1998-08-04 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NH2-terminal sequence truncation decreases the stability of bovine rhodanese, minimally perturbs its crystal structure, and enhances interaction with GroEL under native conditions.
J.Biol.Chem., 274, 1999
|
|
1RHS
 
 | | SULFUR-SUBSTITUTED RHODANESE | | Descriptor: | SULFUR-SUBSTITUTED RHODANESE | | Authors: | Zanotti, G, Gliubich, F, Colapietro, M, Barba, L. | | Deposit date: | 1997-07-16 | | Release date: | 1998-01-21 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structure of sulfur-substituted rhodanese at 1.36 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
6D62
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase I142P from Cupriavidus metallidurans in complex with 3-HAA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXYANTHRANILIC ACID, 3-hydroxyanthranilate 3,4-dioxygenase, ... | | Authors: | Yang, Y, Liu, F, Liu, A. | | Deposit date: | 2018-04-19 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
6D60
 
 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase I142P from Cupriavidus metallidurans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | Authors: | Yang, Y, Liu, F, Liu, A. | | Deposit date: | 2018-04-19 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
4X80
 
 | | Crystal Structure of murine 7B4 Fab monoclonal antibody against ADAMTS5 | | Descriptor: | IgG1 7B4 FAB Heavy chain, IgG1 7B4 FAB Light Chain | | Authors: | Larkin, J, Lohr, T.A, Elefante, L, Shearin, J, Matico, R, Su, J.-L, Xue, Y, Liu, F, Genell, C, Miller, R.E, Tran, P.B, Malfait, A.-M, Maier, C.C, Matheny, C.J. | | Deposit date: | 2014-12-09 | | Release date: | 2015-04-08 | | Last modified: | 2015-08-05 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Translational development of an ADAMTS-5 antibody for osteoarthritis disease modification.
Osteoarthr. Cartil., 23, 2015
|
|
4X8J
 
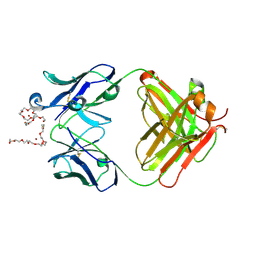 | | Crystal Structure of murine 12F4 Fab monoclonal antibody against ADAMTS5 | | Descriptor: | 12F4 FAB Heavy chain, 12F4 FAB Light chain, NONAETHYLENE GLYCOL, ... | | Authors: | Larkin, J, Lohr, T.A, Elefante, L, Shearin, J, Matico, R, Su, J.-L, Xue, Y, Liu, F, Genell, C, Miller, R.E, Tran, P.B, Malfait, A.-M, Maier, C.C, Matheny, C.J. | | Deposit date: | 2014-12-10 | | Release date: | 2015-04-08 | | Last modified: | 2015-08-05 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Translational development of an ADAMTS-5 antibody for osteoarthritis disease modification.
Osteoarthr. Cartil., 23, 2015
|
|
3FE5
 
 | | Crystal structure of 3-hydroxyanthranilate 3,4-dioxygenase from bovine kidney | | Descriptor: | 3-hydroxyanthranilate 3,4-dioxygenase, FE (III) ION | | Authors: | Dilovic, I, Gliubich, F, Malpeli, G, Zanotti, G, Matkovic-Calogovic, D. | | Deposit date: | 2008-11-27 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structure of bovine 3-hydroxyanthranilate 3,4-dioxygenase.
Biopolymers, 2009
|
|
7BTF
 
 | | SARS-CoV-2 RNA-dependent RNA polymerase in complex with cofactors in reduced condition | | Descriptor: | Non-structural protein 7, Non-structural protein 8, RNA-directed RNA polymerase, ... | | Authors: | Gao, Y, Yan, L, Huang, Y, Liu, F, Cao, L, Wang, T, Wang, Q, Lou, Z, Rao, Z. | | Deposit date: | 2020-04-01 | | Release date: | 2020-04-08 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structure of the RNA-dependent RNA polymerase from COVID-19 virus.
Science, 368, 2020
|
|
6D61
 
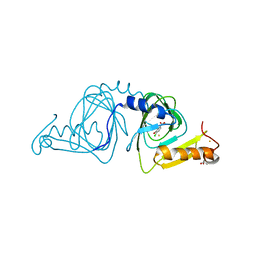 | | Crystal structure of 3-hydroxyanthranilate-3,4-dioxygenase I142P from Cupriavidus metallidurans in complex with 4-Cl-3-HAA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, 4-CHLORO-3-HYDROXYANTHRANILIC ACID, ... | | Authors: | Yang, Y, Liu, F, Liu, A. | | Deposit date: | 2018-04-19 | | Release date: | 2018-06-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Adapting to oxygen: 3-Hydroxyanthrinilate 3,4-dioxygenase employs loop dynamics to accommodate two substrates with disparate polarities.
J. Biol. Chem., 293, 2018
|
|
3WFV
 
 | | HIV-1 CRF07 gp41 | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Du, J, Xue, H, Ma, J, Liu, F, Zhou, J, Shao, Y, Qiao, W, Liu, X. | | Deposit date: | 2013-07-24 | | Release date: | 2013-10-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of HIV CRF07 B'/C gp41 reveals a hyper-mutant site in the middle of HR2 heptad repeat
Virology, 446, 2013
|
|
3NSN
 
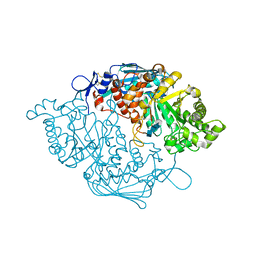 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with TMG-chitotriomycin | | Descriptor: | 2-deoxy-2-(trimethylammonio)-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetylglucosaminidase | | Authors: | Zhang, H, Liu, T, Liu, F, Yang, Q, Shen, X. | | Deposit date: | 2010-07-02 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Determinants of an Insect {beta}-N-Acetyl-D-hexosaminidase Specialized as a Chitinolytic Enzyme
J.Biol.Chem., 286, 2011
|
|
3OZO
 
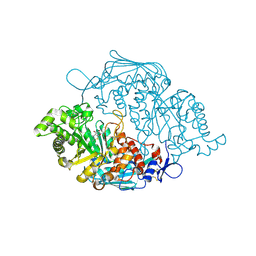 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with NGT | | Descriptor: | 3AR,5R,6S,7R,7AR-5-HYDROXYMETHYL-2-METHYL-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D]THIAZOLE-6,7-DIOL, N-acetylglucosaminidase | | Authors: | Zhang, H, Liu, T, Liu, F, Shen, X, Yang, Q. | | Deposit date: | 2010-09-27 | | Release date: | 2011-09-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with NGT
To be Published
|
|
3OZP
 
 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with PUGNAc | | Descriptor: | N-acetylglucosaminidase, O-(2-ACETAMIDO-2-DEOXY D-GLUCOPYRANOSYLIDENE) AMINO-N-PHENYLCARBAMATE | | Authors: | Zhang, H, Liu, T, Liu, F, Shen, X, Yang, Q. | | Deposit date: | 2010-09-27 | | Release date: | 2011-08-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Active-pocket size differentiating insectile from bacterial chitinolytic beta-N-acetyl-D-hexosaminidases
Biochem.J., 438, 2011
|
|
3NSM
 
 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 from Ostrinia furnacalis | | Descriptor: | N-acetylglucosaminidase | | Authors: | Zhang, H, Liu, T, Liu, F, Yang, Q, Shen, X. | | Deposit date: | 2010-07-02 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Determinants of an Insect {beta}-N-Acetyl-D-hexosaminidase Specialized as a Chitinolytic Enzyme
J.Biol.Chem., 286, 2011
|
|
1XAX
 
 | | NMR structure of HI0004, a putative essential gene product from Haemophilus influenzae | | Descriptor: | Hypothetical UPF0054 protein HI0004 | | Authors: | Yeh, D.C, Parsons, J.F, Parsons, L.M, Liu, F, Eisenstein, E, Orban, J, Structure 2 Function Project (S2F) | | Deposit date: | 2004-08-26 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of HI0004, a putative essential gene product from Haemophilus influenzae, and comparison with the X-ray structure of an Aquifex aeolicus homolog
Protein Sci., 14, 2005
|
|
5V27
 
 | | 2.35 angstrom crystal structure of P97V 3-hydroxyanthranilate-3,4-dioxygenase from Cupriavidus metallidurans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | Authors: | Dornevil, K, Liu, F, Liu, A. | | Deposit date: | 2017-03-02 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | 2.35 angstrom crystal structure of P97V 3-hydroxyanthranilate-3,4-dioxygenase from Cupriavidus metallidurans
To Be Published
|
|
8KG5
 
 | | Prefusion RSV F Bound to Lonafarnib and D25 Fab | | Descriptor: | 4-{2-[4-(3,10-DIBROMO-8-CHLORO-6,11-DIHYDRO-5H-BENZO[5,6]CYCLOHEPTA[1,2-B]PYRIDIN-11-YL)PIPERIDIN-1-YL]-2-OXOETHYL}PIPERIDINE-1-CARBOXAMIDE, D25 heavy chain, D25 light chain, ... | | Authors: | Yang, Q, Xue, B, Liu, F, Peng, W, Chen, X. | | Deposit date: | 2023-08-17 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Farnesyltransferase inhibitor lonafarnib suppresses respiratory syncytial virus infection by blocking conformational change of fusion glycoprotein.
Signal Transduct Target Ther, 9, 2024
|
|
5V28
 
 | | 2.72 angstrom crystal structure of P97A 3-hydroxyanthranilate-3,4-dioxygenase from Cupriavidus metallidurans | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-hydroxyanthranilate 3,4-dioxygenase, FE (II) ION | | Authors: | Dornevil, K, Liu, F, Liu, A. | | Deposit date: | 2017-03-02 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.724 Å) | | Cite: | 2.72 angstrom crystal structure of P97A 3-hydroxyanthranilate-3,4-dioxygenase from Cupriavidus metallidurans
To Be Published
|
|
