3FDE
 
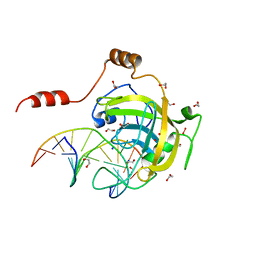 | | Mouse UHRF1 SRA domain bound with hemi-methylated CpG DNA, crystal structure in space group C222(1) at 1.4 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*CP*CP*AP*TP*GP*(5CM)P*GP*CP*TP*GP*AP*C)-3', 5'-D(*GP*TP*CP*AP*GP*CP*GP*CP*AP*TP*GP*G)-3', ... | | Authors: | Hashimoto, H, Horton, J.R, Zhang, X, Cheng, X. | | Deposit date: | 2008-11-25 | | Release date: | 2009-01-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications.
Epigenetics, 4, 2009
|
|
3F8J
 
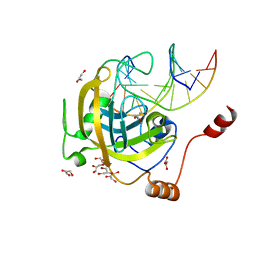 | | Mouse UHRF1 SRA domain bound with hemi-methylated CpG, crystal structure in space group C222(1) | | Descriptor: | 5'-D(*DCP*DCP*DAP*DTP*DGP*(5CM)P*DGP*DCP*DTP*DGP*DAP*DC)-3', 5'-D(*DGP*DTP*DCP*DAP*DGP*DCP*DGP*DCP*DAP*DTP*DGP*DG)-3', E3 ubiquitin-protein ligase UHRF1, ... | | Authors: | Hashimoto, H, Horton, J.R, Zhang, X, Cheng, X. | | Deposit date: | 2008-11-12 | | Release date: | 2009-01-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications.
Epigenetics, 4, 2009
|
|
5IAY
 
 | | NMR structure of UHRF1 Tandem Tudor Domains in a complex with Spacer peptide | | Descriptor: | E3 ubiquitin-protein ligase UHRF1, Spacer | | Authors: | Fang, J, Cheng, J, Wang, J, Zhang, Q, Liu, M, Gong, R, Wang, P, Zhang, X, Feng, Y, Lan, W, Gong, Z, Tang, C, Wong, J, Yang, H, Cao, C, Xu, Y. | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Hemi-methylated DNA opens a closed conformation of UHRF1 to facilitate its histone recognition
Nat Commun, 7, 2016
|
|
2XDN
 
 | |
5J6P
 
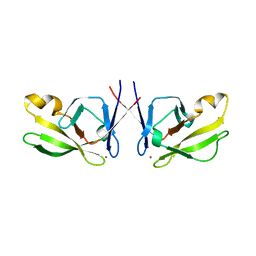 | | Crystal Structure of Mis18(17-118) from Schizosaccharomyces pombe | | Descriptor: | Kinetochore protein mis18, ZINC ION | | Authors: | Wang, C, Shao, C, Zhang, M, Zhang, X, Zang, J. | | Deposit date: | 2016-04-05 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Mis18(17-118) from Schizosaccharomyces pombe
To Be Published
|
|
8KGF
 
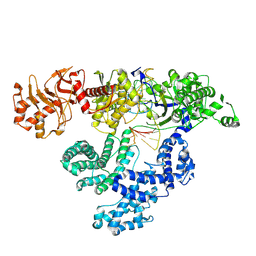 | | Structure of AmCas12a with crRNA | | Descriptor: | CRISPR-associated endonuclease Cas12a, MAGNESIUM ION, RNA (44-MER) | | Authors: | Feng, Y, Zhang, X, Shi, J, Ma, P, Tang, J, Huang, X. | | Deposit date: | 2023-08-18 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A novel CRISPR/Cas12a in complex with a crRNA.
To Be Published
|
|
2XRO
 
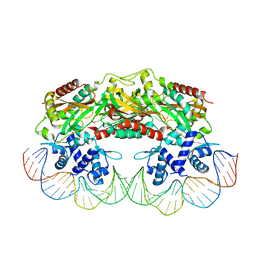 | | Crystal structure of TtgV in complex with its DNA operator | | Descriptor: | HTH-TYPE TRANSCRIPTIONAL REGULATOR TTGV, OSMIUM ION, TTGV OPERATOR DNA | | Authors: | Lu, D, Fillet, S, Meng, C, Alguel, Y, Kloppsteck, P, Bergeron, J, Krell, T, Gallegos, M.-T, Ramos, J, Zhang, X. | | Deposit date: | 2010-09-17 | | Release date: | 2010-12-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of Ttgv in Complex with its DNA Operator Reveals a General Model for Cooperative DNA Binding of Tetrameric Gene Regulators.
Genes Dev., 24, 2010
|
|
2XRN
 
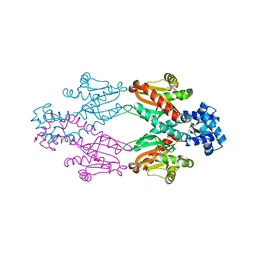 | | Crystal structure of TtgV | | Descriptor: | HTH-TYPE TRANSCRIPTIONAL REGULATOR TTGV | | Authors: | Lu, D, Fillet, S, Meng, C, Alguel, Y, Kloppsteck, P, Bergeron, J, Krell, T, Gallegos, M.-T, Ramos, J, Zhang, X. | | Deposit date: | 2010-09-17 | | Release date: | 2010-12-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal Structure of Ttgv in Complex with its DNA Operator Reveals a General Model for Cooperative DNA Binding of Tetrameric Gene Regulators.
Genes Dev., 24, 2010
|
|
7NTS
 
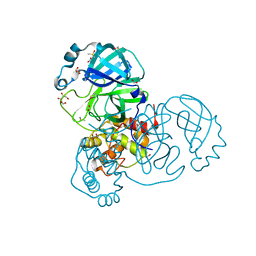 | | Crystal structure of the SARS-CoV-2 Main Protease with oxidized C145 | | Descriptor: | DIMETHYL SULFOXIDE, FORMIC ACID, GLYCEROL, ... | | Authors: | Dupre, E, Villeret, V, Hanoulle, X. | | Deposit date: | 2021-03-10 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.477 Å) | | Cite: | NMR Spectroscopy of the Main Protease of SARS-CoV-2 and Fragment-Based Screening Identify Three Protein Hotspots and an Antiviral Fragment.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
5ABF
 
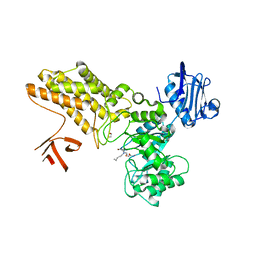 | | Structure of GH84 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2S,3R,4R,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)-1-pentyl-pyrrolidin-2-yl]-N-methyl-ethanamide, CALCIUM ION, ... | | Authors: | Bergeron-Brlek, M, Goodwin-Tindall, J, Cekic, N, Varghese, V, Zandberg, W.F, Shan, X, Roth, C, Chan, S, Davies, G.J, Vocadlo, D.J, Britton, R. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Convenient Approach to Stereoisomeric Iminocyclitols: Generation of Potent Brain-Permeable Oga Inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
1G55
 
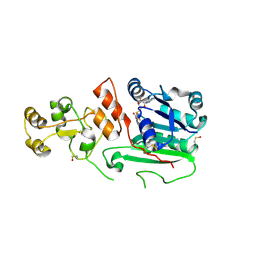 | | Structure of human DNMT2, an enigmatic DNA methyltransferase homologue | | Descriptor: | BETA-MERCAPTOETHANOL, DNA CYTOSINE METHYLTRANSFERASE DNMT2, GLYCEROL, ... | | Authors: | Dong, A, Yoder, J.A, Zhang, X, Zhou, L, Bestor, T.H, Cheng, X. | | Deposit date: | 2000-10-30 | | Release date: | 2001-01-17 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human DNMT2, an enigmatic DNA methyltransferase homolog that displays denaturant-resistant binding to DNA.
Nucleic Acids Res., 29, 2001
|
|
7NTQ
 
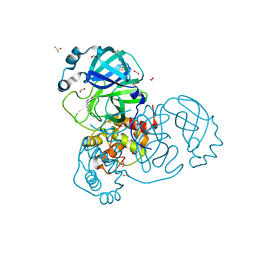 | | Crystal structure of the SARS-CoV-2 Main Protease complexed with N-(pyridin-3-ylmethyl)thioformamide | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Dupre, E, Villeret, V, Hanoulle, X. | | Deposit date: | 2021-03-10 | | Release date: | 2022-03-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.495 Å) | | Cite: | Novel dithiocarbamates selectively inhibit 3CL protease of SARS-CoV-2 and other coronaviruses.
Eur.J.Med.Chem., 250, 2023
|
|
7NTW
 
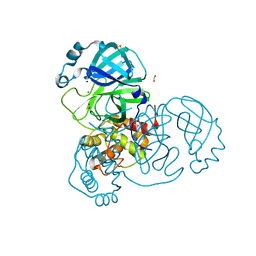 | |
7NTT
 
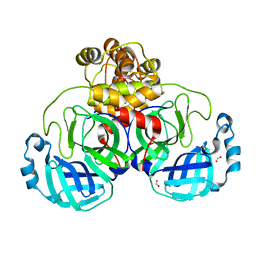 | |
1P9E
 
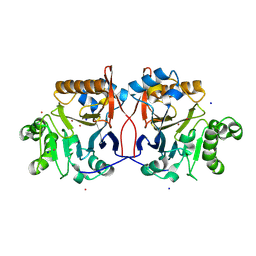 | | Crystal Structure Analysis of Methyl Parathion Hydrolase from Pseudomonas sp WBC-3 | | Descriptor: | CADMIUM ION, Methyl Parathion Hydrolase, POTASSIUM ION, ... | | Authors: | Dong, Y, Sun, L, Bartlam, M, Rao, Z, Zhang, X. | | Deposit date: | 2003-05-11 | | Release date: | 2004-05-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure Analysis of Methyl Parathion Hydrolase from Pseudomonas sp WBC-3
To be Published
|
|
7Y7A
 
 | | In situ double-PBS-PSII-PSI-LHCs megacomplex from Porphyridium purpureum. | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sun, S, Sui, S.F. | | Deposit date: | 2022-06-22 | | Release date: | 2023-02-08 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
7Y5E
 
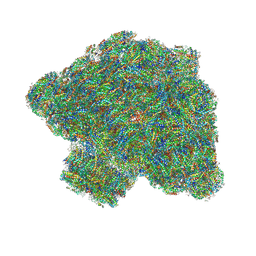 | | In situ single-PBS-PSII-PSI-LHCs megacomplex. | | Descriptor: | (1R,2S)-4-{(1E,3E,5E,7E,9E,11E,13E,15E,17E)-18-[(4S)-4-hydroxy-2,6,6-trimethylcyclohex-1-en-1-yl]-3,7,12,16-tetramethyloctadeca-1,3,5,7,9,11,13,15,17-nonaen-1-yl}-2,5,5-trimethylcyclohex-3-en-1-ol, (2S)-2,3-dihydroxypropyl octadecanoate, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | You, X, Zhang, X, Cheng, J, Xiao, Y.N, Sui, S.F. | | Deposit date: | 2022-06-17 | | Release date: | 2023-02-01 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | In situ structure of the red algal phycobilisome-PSII-PSI-LHC megacomplex.
Nature, 616, 2023
|
|
5IHW
 
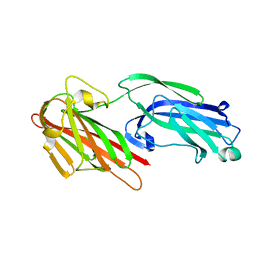 | | The crystal structure of SdrE from staphylococcus aureus | | Descriptor: | Serine-aspartate repeat-containing protein E | | Authors: | Zhang, S, Wei, J, Wu, S, Zhang, X, Luo, M, Wang, D. | | Deposit date: | 2016-02-29 | | Release date: | 2017-03-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | The crystal structure of SdrE from staphylococcus aureus
To Be Published
|
|
1SVU
 
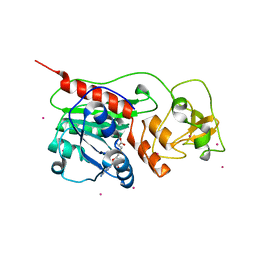 | | Structure of the Q237W mutant of HhaI DNA methyltransferase: an insight into protein-protein interactions | | Descriptor: | Modification methylase HhaI, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Dong, A, Zhou, L, Zhang, X, Stickel, S, Roberts, R.J, Cheng, X. | | Deposit date: | 2004-03-30 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structure of the Q237W mutant of HhaI DNA methyltransferase: an insight into protein-protein interactions
Biol.Chem., 385, 2004
|
|
1VAZ
 
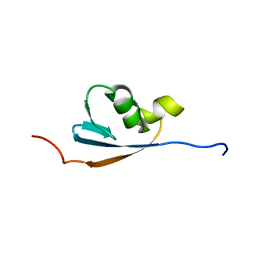 | | Solution structures of the p47 SEP domain | | Descriptor: | NSFL1 cofactor p47 | | Authors: | Yuan, X, Simpson, P, Mckeown, C, Kondo, H, Uchiyama, K, Wallis, R, Dreveny, I, Keetch, C, Zhang, X, Robinson, C, Freemont, P, Matthews, S. | | Deposit date: | 2004-02-20 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics and interactions of p47, a major adaptor of the AAA ATPase, p97.
Embo J., 23, 2004
|
|
1V92
 
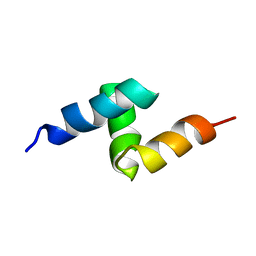 | | Solution structure of the UBA domain from p47, a major cofactor of the AAA ATPase p97 | | Descriptor: | NSFL1 cofactor p47 | | Authors: | Yuan, X, Simpson, P, Mckeown, C, Kondo, H, Uchiyama, K, Wallis, R, Dreveny, I, Keetch, C, Zhang, X, Robinson, C, Freemont, P, Matthews, S. | | Deposit date: | 2004-01-19 | | Release date: | 2004-04-06 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics and interactions of p47, a major adaptor of the AAA ATPase, p97
Embo J., 23, 2004
|
|
1U2Z
 
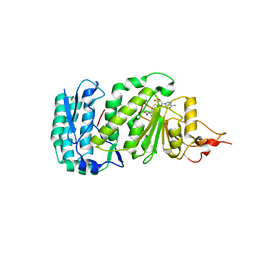 | | Crystal structure of histone K79 methyltransferase Dot1p from yeast | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Sawada, K, Yang, Z, Horton, J.R, Collins, R.E, Zhang, X, Cheng, X. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the conserved core of the yeast Dot1p, a nucleosomal histone H3 lysine 79 methyltransferase
J.Biol.Chem., 279, 2004
|
|
5EWU
 
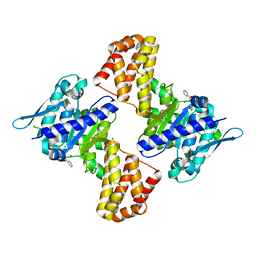 | | Crystal structure of the Arabidopsis thaliana C-terminal Chlh at 1.25A | | Descriptor: | BENZOIC ACID, MAGNESIUM ION, Magnesium-chelatase subunit ChlH, ... | | Authors: | Chen, Z, Zhang, X, Liu, Y, Jiang, L. | | Deposit date: | 2015-11-21 | | Release date: | 2016-12-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of the Arabidopsis thaliana C-terminal Chlh at 1.25A
To Be Published
|
|
1RKU
 
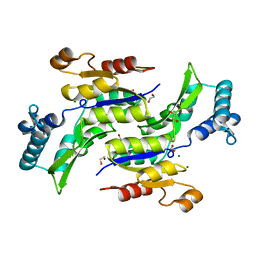 | | Crystal Structure of ThrH gene product of Pseudomonas Aeruginosa | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, homoserine kinase | | Authors: | Singh, S.K, Yang, K, Subramanian, K, Karthikeyan, S, Huynh, T, Zhang, X, Phillips, M.A, Zhang, H. | | Deposit date: | 2003-11-23 | | Release date: | 2004-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The thrH Gene Product of Pseudomonas aeruginosa Is a Dual Activity Enzyme with a Novel Phosphoserine:Homoserine Phosphotransferase Activity.
J.Biol.Chem., 279, 2004
|
|
6LS3
 
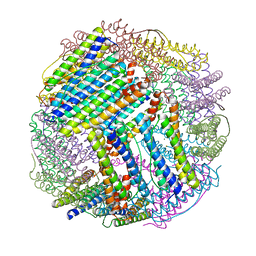 | |
