7APE
 
 | | Crystal structure of LpqY from Mycobacterium thermoresistible in complex with trehalose | | Descriptor: | Lipoprotein (Sugar-binding) lpqY, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Furze, C.M, Guy, C.M, Angula, J, Cameron, A.D, Fullam, E. | | Deposit date: | 2020-10-16 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of trehalose recognition by the mycobacterial LpqY-SugABC transporter.
J.Biol.Chem., 296, 2021
|
|
3S7F
 
 | |
3QUL
 
 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with LCMV-derived gp33 altered peptide ligand (Y4S) | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Allerbring, E, Duru, A.D, Uchtenhagen, H, Madhurantakam, C, Grimm, S, Tomek, M.B, Mazumdar, P.A, Spetz, A, Friemann, R, Sandalova, T, Uhlin, M, Nygren, P, Achour, A. | | Deposit date: | 2011-02-24 | | Release date: | 2012-03-21 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unexpected T-cell recognition of an altered peptide ligand is driven by reversed thermodynamics.
Eur.J.Immunol., 42, 2012
|
|
8UND
 
 | | X-ray Structure of SARS-CoV-2 main protease covalently bound to inhibitor GRL-190-21 at 1.90 A. | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopiperidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, ORF1a polyprotein | | Authors: | Mesecar, A.D, Lendy, E.K, Ghosh, A.K, Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-10-18 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray Structure of SARS-CoV-2 main protease covalently bound to inhibitor GRL-190-21 at 1.90 A.
To Be Published
|
|
8UM5
 
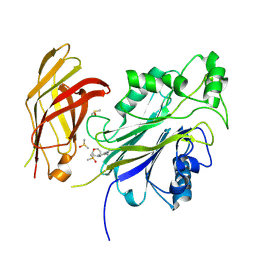 | |
3S26
 
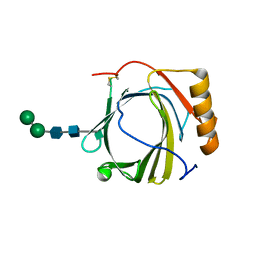 | | Crystal Structure of Murine Siderocalin (Lipocalin 2, 24p3) | | Descriptor: | Neutrophil gelatinase-associated lipocalin, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Correnti, C, Bandaranayake, A.D, Strong, R.K. | | Deposit date: | 2011-05-16 | | Release date: | 2011-09-28 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Daedalus: a robust, turnkey platform for rapid production of decigram quantities of active recombinant proteins in human cell lines using novel lentiviral vectors.
Nucleic Acids Res., 39, 2011
|
|
7PI6
 
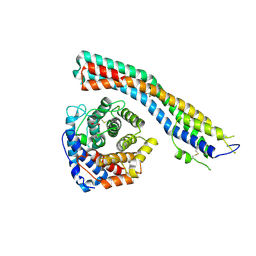 | | Trypanosoma brucei ISG65 bound to human complement C3d | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 65 kDa invariant surface glycoprotein, Complement C3dg fragment, ... | | Authors: | Cook, A.D, Higgins, M.K. | | Deposit date: | 2021-08-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Invariant surface glycoprotein 65 of Trypanosoma brucei is a complement C3 receptor.
Nat Commun, 13, 2022
|
|
7AEA
 
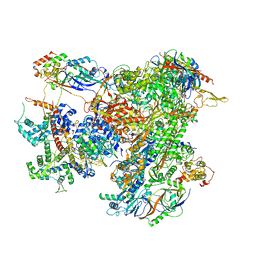 | | Cryo-EM structure of human RNA Polymerase III elongation complex 2 | | Descriptor: | DNA-directed RNA polymerase III subunit RPC1, DNA-directed RNA polymerase III subunit RPC10, DNA-directed RNA polymerase III subunit RPC2, ... | | Authors: | Girbig, M, Misiaszek, A.D, Vorlaender, M.K, Mueller, C.W. | | Deposit date: | 2020-09-17 | | Release date: | 2021-02-03 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of human RNA polymerase III in its unbound and transcribing states.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7RQ5
 
 | |
7QER
 
 | | human Connexin 26 dodecamer at 55mm Hg PCO2, pH7.4 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QEQ
 
 | | human Connexin 26 dodecamer at 90mmHg PCO2, pH7.4 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
8VKZ
 
 | | Crystal structure of Glucocorticoid Receptor in complex with an inhibitor | | Descriptor: | (4aR,4bS,5R,6aS,6bS,8R,9aR,10aR,10bR)-8-{4-[(3-aminophenyl)methyl]phenyl}-5-hydroxy-6b-(hydroxyacetyl)-4a,6a-dimethyl-4a,4b,5,6,6a,6b,9a,10,10a,10b,11,12-dodecahydro-2H,8H-naphtho[2',1':4,5]indeno[1,2-d][1,3]dioxol-2-one, Glucocorticoid receptor, Nuclear receptor coactivator 2 | | Authors: | Judge, R.A, Hobson, A.D. | | Deposit date: | 2024-01-10 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | Minimising the payload solvent exposed hydrophobic surface area optimises the antibody-drug conjugate properties.
Rsc Med Chem, 15, 2024
|
|
7QET
 
 | | human Connexin 26 dodecamer at 20mmHg PCO2, pH7.4 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-03-30 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QEW
 
 | | human Connexin 26 class 2 hexamer at 90mmHg PCO2, pH7.4 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-03-30 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
7QEY
 
 | | human Connexin 26 class 1 hexamer at 90mmHg PCO2, pH7.4 | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, Gap junction beta-2 protein, PHOSPHATIDYLETHANOLAMINE | | Authors: | Brotherton, D.H, Cameron, A.D, Savva, C.G, Ragan, T.J. | | Deposit date: | 2021-12-03 | | Release date: | 2022-03-30 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Conformational changes and CO 2 -induced channel gating in connexin26.
Structure, 30, 2022
|
|
3G5M
 
 | | Synthesis of Casimiroin and Optimization of Its Quinone Reductase 2 and Aromatase Inhibitory activity | | Descriptor: | 6-methoxy-9-methyl[1,3]dioxolo[4,5-h]quinolin-8(9H)-one, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Maiti, A, Sturdy, M, Marler, L, Pegan, S.D, Mesecar, A.D, Pezzuto, J.M, Cushman, M. | | Deposit date: | 2009-02-05 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Synthesis of casimiroin and optimization of its quinone reductase 2 and aromatase inhibitory activities.
J.Med.Chem., 52, 2009
|
|
3GAM
 
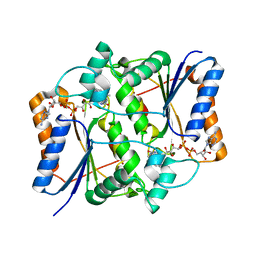 | | Synthesis of Casimiroin and Optimization of Its Quinone Reductase 2 and Aromatase Inhibitory activity | | Descriptor: | 5,8-dimethoxy-1,4-dimethylquinolin-2(1H)-one, FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ... | | Authors: | Sturdy, M, Pegan, S.D, Maiti, A, Marler, L, Mesecar, A.D, Pezzuto, J.M, Cushman, M. | | Deposit date: | 2009-02-17 | | Release date: | 2009-03-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Synthesis of casimiroin and optimization of its quinone reductase 2 and aromatase inhibitory activities.
J.Med.Chem., 52, 2009
|
|
3TBT
 
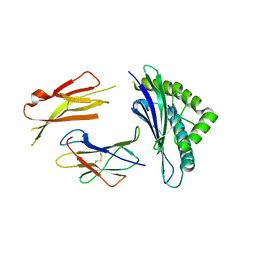 | | CRYSTAL STRUCTURE OF THE MURINE CLASS I MAJOR HISTOCOMPATIBILITY COMPLEX H-2DB IN COMPLEX WITH THE LCMV-DERIVED GP33 ALTERED PEPTIDE ligand (V3P, Y4S) | | Descriptor: | Beta-2-microglobulin, GLYCOPROTEIN G1, H-2 class I histocompatibility antigen, ... | | Authors: | Duru, A.D, Allerbring, E.B, Uchtenhagen, H, Mazumdar, P.A, Badia-Martinez, D, Madhurantakam, C, Sandalova, T, Nygren, P, Achour, A. | | Deposit date: | 2011-08-08 | | Release date: | 2012-08-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Conversion of a T cell viral antagonist into an agonist through higher stabilization and conserved molecular mimicry: Implications for TCR recognition
To be Published
|
|
3G5E
 
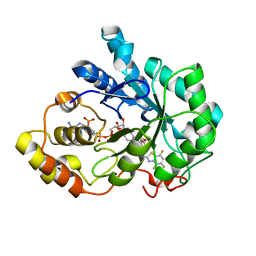 | | Human aldose reductase complexed with IDD 740 inhibitor | | Descriptor: | 2-(3-((4,5,7-trifluorobenzo[d]thiazol-2-yl)methyl)-1H-pyrrolo[2,3-b]pyridin-1-yl)acetic acid, Aldose reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Podjarny, A.D, Van Zandt, M.C. | | Deposit date: | 2009-02-05 | | Release date: | 2009-03-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of [3-(4,5,7-trifluoro-benzothiazol-2-ylmethyl)-pyrrolo[2,3-b]pyridin-1-yl]acetic acids as highly potent and selective inhibitors of aldose reductase for treatment of chronic diabetic complications.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3TBV
 
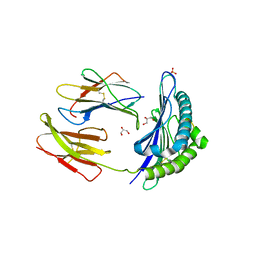 | | CRYSTAL STRUCTURE OF THE MURINE CLASS I MAJOR HISTOCOMPATIBILITY COMPLEX H-2DB IN COMPLEX WITH THE LCMV-DERIVED GP33 ALTERED PEPTIDE ligand (A2G,V3P,Y4A) | | Descriptor: | Beta-2-microglobulin, GLYCEROL, Glycoprotein G1, ... | | Authors: | Duru, A.D, Allerbring, E.B, Uchtenhagen, H, Mazumdar, P.A, Badia-Martinez, D, Madhurantakam, C, Sandalova, T, Nygren, P, Achour, A. | | Deposit date: | 2011-08-08 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conversion of a T cell viral antagonist into an agonist through higher stabilization and conserved molecular mimicry: Implications for TCR recognition
To be Published
|
|
2LS5
 
 | | Solution structure of a putative protein disulfide isomerase from Bacteroides thetaiotaomicron | | Descriptor: | Uncharacterized protein | | Authors: | Harris, R, Bandaranayake, A.D, Banu, R, Bonanno, J.B, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chaparro, R, Evans, B, Garforth, S, Gizzi, A, Hillerich, B, Kar, A, Lafleur, J, Lim, S, Love, J, Matikainen, B, Patel, H, Seidel, R.D, Smith, B, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-04-20 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a putative protein disulfide isomerase from Bacteroides thetaiotaomicron
To be Published
|
|
3TBW
 
 | | CRYSTAL STRUCTURE OF THE MURINE CLASS I MAJOR HISTOCOMPATIBILITY COMPLEX H-2DB IN COMPLEX WITH THE LCMV-DERIVED GP33 ALTERED PEPTIDE ligand (A2G, V3P, Y4S) | | Descriptor: | Beta-2-microglobulin, GLYCOPROTEIN GPC, H-2 class I histocompatibility antigen, ... | | Authors: | Duru, A.D, Allerbring, E.B, Uchtenhagen, H, Mazumdar, P.A, Badia-Martinez, D, Madhurantakam, C, Sandalova, T, Nygren, P, Achour, A. | | Deposit date: | 2011-08-08 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conversion of a T cell viral antagonist into an agonist through higher stabilization and conserved molecular mimicry: Implications for TCR recognition
To be Published
|
|
2GZM
 
 | | Crystal Structure of the Glutamate Racemase from Bacillus anthracis | | Descriptor: | D-GLUTAMIC ACID, Glutamate racemase | | Authors: | May, M, Santarsiero, B.D, Johnson, M.E, Mesecar, A.D. | | Deposit date: | 2006-05-11 | | Release date: | 2007-05-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and functional analysis of two glutamate racemase isozymes from Bacillus anthracis and implications for inhibitor design.
J.Mol.Biol., 371, 2007
|
|
3DMT
 
 | | Structure of Glycosomal Glyceraldehyde-3-Phosphate Dehydrogenase from Trypanosoma cruzi in complex with the irreversible iodoacetate inhibitor | | Descriptor: | GLYCEROL, Glyceraldehyde-3-phosphate dehydrogenase, glycosomal, ... | | Authors: | Guido, R.V.C, Balliano, T.L, Andricopulo, A.D, Oliva, G. | | Deposit date: | 2008-07-01 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and Crystallographic Studies on Glyceraldehyde-3-Phosphate Dehydrogenase from Trypanosoma cruzi in Complex with Iodoacetate.
Letters in drug design & discovery, 6, 2009
|
|
3GE8
 
 | | Toluene 4-monooxygenase HD T201A diferric, resting state complex | | Descriptor: | ACETATE ION, FE (III) ION, Toluene-4-monooxygenase system protein A, ... | | Authors: | Elsen, N.L, Bailey, L.J, Hauser, A.D, Fox, B.G. | | Deposit date: | 2009-02-25 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Role for threonine 201 in the catalytic cycle of the soluble diiron hydroxylase toluene 4-monooxygenase.
Biochemistry, 48, 2009
|
|
