4TY7
 
 | |
4TY6
 
 | |
8X80
 
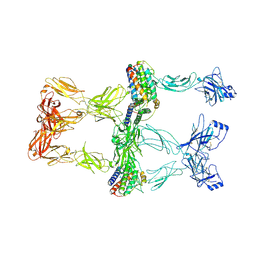 | | Structure of leptin-LepR trimer with a small gap | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Leptin, ... | | Authors: | Xie, Y.F, Gao, G.F. | | Deposit date: | 2023-11-27 | | Release date: | 2024-08-07 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Structural plasticity of human leptin binding to its receptor LepR
Hlife, 1, 2023
|
|
8X81
 
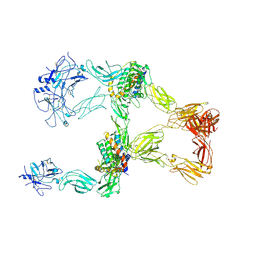 | | Structure of leptin-LepR trimer with a large gap | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Leptin, ... | | Authors: | Xie, Y.F, Gao, G.F. | | Deposit date: | 2023-11-27 | | Release date: | 2024-08-07 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Structural plasticity of human leptin binding to its receptor LepR
Hlife, 1, 2023
|
|
6LMJ
 
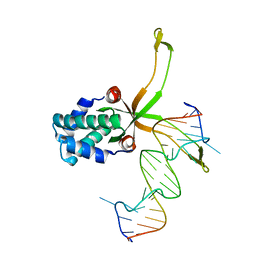 | | ASFV pA104R in complex with double-strand DNA | | Descriptor: | A104R, DNA (5'-D(*TP*GP*CP*TP*TP*AP*TP*CP*AP*AP*TP*TP*TP*GP*TP*TP*GP*CP*A)-3') | | Authors: | Wang, H, Qi, J, Chai, Y, Gao, F, Liu, R. | | Deposit date: | 2019-12-25 | | Release date: | 2020-05-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis of African swine fever virus pA104R binding to DNA and its inhibition by stilbene derivatives.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6LMH
 
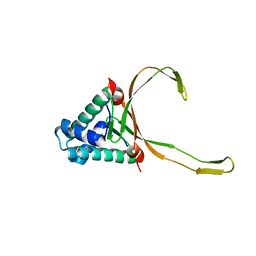 | |
6ZW1
 
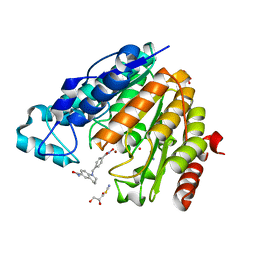 | | X-ray structure of Danio rerio histone deacetylase 6 (HDAC6) CD2 in complex with an inhibitor SW101 | | Descriptor: | 1-[[4-(oxidanylcarbamoyl)phenyl]methyl]-3,4-dihydro-2~{H}-quinoline-6-carboxamide, GLYCEROL, Histone deacetylase 6, ... | | Authors: | Barinka, C, Motlova, L, Ustinova, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Tetrahydroquinoline-Capped Histone Deacetylase 6 Inhibitor SW-101 Ameliorates Pathological Phenotypes in a Charcot-Marie-Tooth Type 2A Mouse Model.
J.Med.Chem., 64, 2021
|
|
6MTA
 
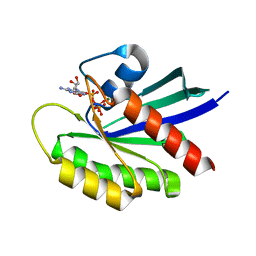 | |
6A06
 
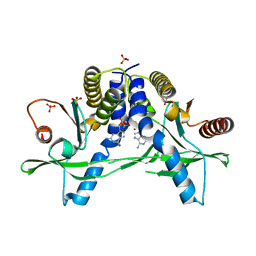 | | Structure of pSTING complex | | Descriptor: | SULFATE ION, Stimulator of interferon genes protein, cGAMP | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6A04
 
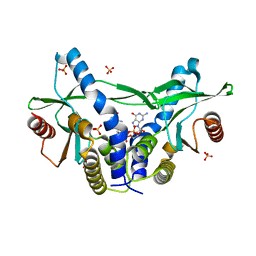 | | Structure of pSTING complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), SULFATE ION, Stimulator of interferon genes protein | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
9E5F
 
 | | Discovery of an Orally Bioavailable KRAS G12D Inhibitor | | Descriptor: | (4P)-4-{1-[(1R,4R,5S)-2-azabicyclo[2.1.1]hexan-5-yl]-8-chloro-4-[3-(dimethylamino)azetidin-1-yl]-6-fluoro-1H-imidazo[4,5-c]quinolin-7-yl}naphthalen-2-ol, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Deller, M.C, Epling, L.B. | | Deposit date: | 2024-10-28 | | Release date: | 2025-01-22 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Discovery of INCB159020, an Orally Bioavailable KRAS G12D Inhibitor.
J.Med.Chem., 68, 2025
|
|
9E5D
 
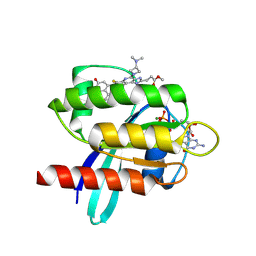 | |
6MS9
 
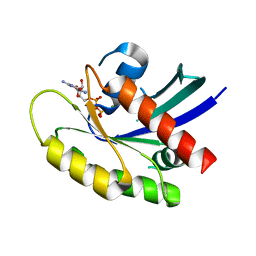 | | GDP-bound KRAS P34R mutant | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Bera, A.K, Westover, K.D. | | Deposit date: | 2018-10-16 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | GTP hydrolysis is modulated by Arg34 in the RASopathy-associated KRASP34R.
Birth Defects Res, 112, 2020
|
|
6A03
 
 | | Structure of pSTING complex | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, SULFATE ION, Stimulator of interferon genes protein | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
6O36
 
 | |
6GY9
 
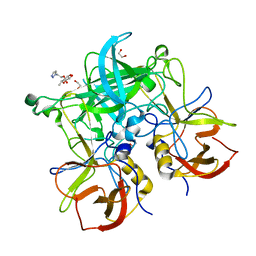 | | Fucose-functionalized precision glycomacromolecules targeting human norovirus capsid protein | | Descriptor: | 1,2-ETHANEDIOL, 2-(1H-1,2,3-triazol-1-yl)ethyl 6-deoxy-alpha-L-galactopyranoside, Capsid protein | | Authors: | Ruoff, K, Kilic, T, Hansman, G.S. | | Deposit date: | 2018-06-28 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Fucose-Functionalized Precision Glycomacromolecules Targeting Human Norovirus Capsid Protein.
Biomacromolecules, 19, 2018
|
|
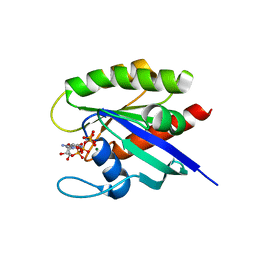
6O46
 
 | |
6H70
 
 | |
6H6Y
 
 | | GI.1 human norovirus protruding domain in complex with Nano-7 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Capsid protein VP1, ... | | Authors: | Kilic, T, Ruoff, K, Hansman, G.S. | | Deposit date: | 2018-07-30 | | Release date: | 2018-12-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Basis of Nanobodies Targeting the Prototype Norovirus.
J. Virol., 93, 2019
|
|
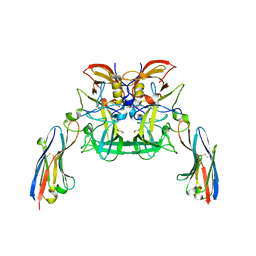
6H71
 
 | |
6H6Z
 
 | |
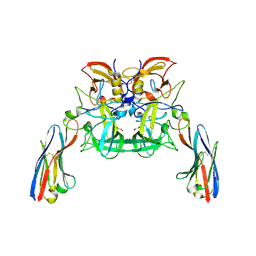
6H72
 
 | |
6A05
 
 | | Structure of pSTING complex | | Descriptor: | 2-amino-9-[(2R,3R,3aR,5S,7aS,9R,10R,10aR,12R,14aS)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, SULFATE ION, Stimulator of interferon genes protein | | Authors: | Yuan, Z.L, Shang, G.J, Cong, X.Y, Gu, L.C. | | Deposit date: | 2018-06-05 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of porcine STINGCBD-CDN complexes reveal the mechanism of ligand recognition and discrimination of STING proteins.
J.Biol.Chem., 294, 2019
|
|
4U0L
 
 | |
4U0N
 
 | | Structure of the Vibrio cholerae di-nucleotide cyclase (DncV) deletion mutant D-loop | | Descriptor: | Cyclic AMP-GMP synthase, MAGNESIUM ION, N-[4-({[(6S)-2-amino-5-methyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-gamma-glutamyl-L-glutamic acid | | Authors: | Xiang, Y, Zhu, D.Y. | | Deposit date: | 2014-07-12 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structural Biochemistry of a Vibrio cholerae Dinucleotide Cyclase Reveals Cyclase Activity Regulation by Folates.
Mol.Cell, 55, 2014
|
|
