7XB0
 
 | | Crystal structure of Omicron BA.2 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L, Liao, H, Meng, Y, Li, W. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
7XAZ
 
 | | Crystal structure of Omicron BA.1.1 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Liao, H, Meng, Y, Li, W. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
7XB1
 
 | | Crystal structure of Omicron BA.3 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, W, Meng, Y, Liao, H. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
6B5Q
 
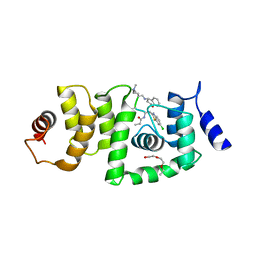 | | DCN1 bound to 38 | | Descriptor: | DCN1-like protein 1, Peptidomimetic Inhibitors DI-591, TRIETHYLENE GLYCOL | | Authors: | Stuckey, J. | | Deposit date: | 2017-09-29 | | Release date: | 2018-02-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | High-Affinity Peptidomimetic Inhibitors of the DCN1-UBC12 Protein-Protein Interaction.
J. Med. Chem., 61, 2018
|
|
2KMZ
 
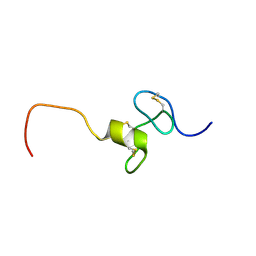 | | NMR Structure of hFn14 | | Descriptor: | Tumor necrosis factor receptor superfamily member 12A | | Authors: | Pellegrini, M, Willen, L, Perroud, M, Krushinskie, D, Strauch, K, Cuervo, H, Sun, Y, Day, E.S, Schneider, P, Zheng, T.S. | | Deposit date: | 2009-08-10 | | Release date: | 2011-06-29 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the extracellular domains of human and Xenopus Fn14: implications in the evolution of TWEAK and Fn14 interactions.
Febs J., 280, 2013
|
|
2KN0
 
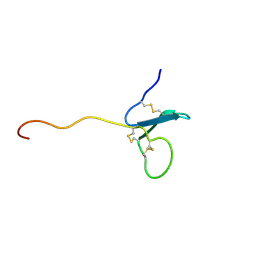 | | Solution NMR Structure of xenopus Fn14 | | Descriptor: | Fn14 | | Authors: | Pellegrini, M, Willen, L, Perroud, M, Krushinskie, D, Strauch, K, Cuervo, H, Sun, Y, Day, E.S, Schneider, P, Zheng, T.S. | | Deposit date: | 2009-08-11 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Structure of the extracellular domains of human and Xenopus Fn14: implications in the evolution of TWEAK and Fn14 interactions.
Febs J., 280, 2013
|
|
8FAQ
 
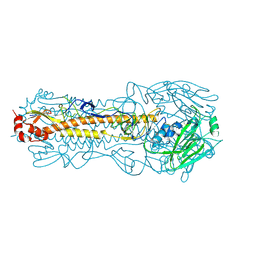 | | Structure of Hemagglutinin from Influenza A/Victoria/22/2020 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Hernandez Garcia, A, Lei, R. | | Deposit date: | 2022-11-28 | | Release date: | 2024-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.035 Å) | | Cite: | Epistasis mediates the evolution of the receptor binding mode in recent human H3N2 hemagglutinin.
Nat Commun, 15, 2024
|
|
8FAW
 
 | |
7YYO
 
 | | Cryo-EM structure of an a-carboxysome RuBisCO enzyme at 2.9 A resolution | | Descriptor: | 2-CARBOXYARABINITOL-1,5-DIPHOSPHATE, MAGNESIUM ION, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Mann, D, Evans, S.L, Bergeron, J.R.C. | | Deposit date: | 2022-02-18 | | Release date: | 2023-01-25 | | Last modified: | 2023-06-14 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Single-particle cryo-EM analysis of the shell architecture and internal organization of an intact alpha-carboxysome.
Structure, 31, 2023
|
|
8XRY
 
 | |
8XNG
 
 | |
8XS4
 
 | |
8XW1
 
 | | Cryo-EM structure of OSCA1.2-V335W-DDM state | | Descriptor: | Calcium permeable stress-gated cation channel 1 | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-04-10 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.49 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
8XW2
 
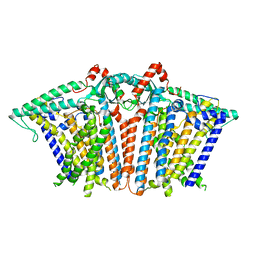 | |
8XS5
 
 | |
8XW3
 
 | |
8XVY
 
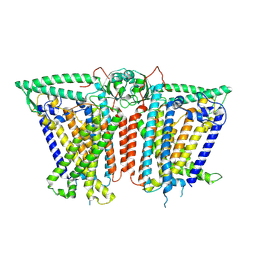 | |
8XW0
 
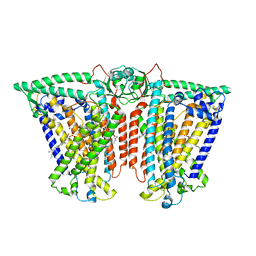 | | Cryo-EM structure of OSCA3.1-GDN state | | Descriptor: | CSC1-like protein ERD4, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-04-10 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
8XVZ
 
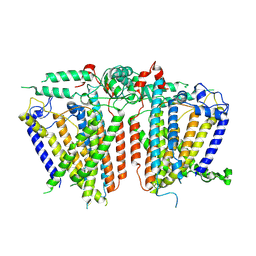 | |
8XW4
 
 | | Cryo-EM structure of TMEM63B-Digitonin state | | Descriptor: | CSC1-like protein 2 | | Authors: | Zhang, Y, Han, Y. | | Deposit date: | 2024-01-15 | | Release date: | 2024-04-10 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Mechanical activation opens a lipid-lined pore in OSCA ion channels.
Nature, 628, 2024
|
|
8XS0
 
 | |
8XVX
 
 | |
8G1U
 
 | | Structure of the methylosome-Lsm10/11 complex | | Descriptor: | ADENOSINE, Methylosome protein 50, Methylosome subunit pICln, ... | | Authors: | Lin, M, Paige, A, Tong, L. | | Deposit date: | 2023-02-03 | | Release date: | 2023-08-23 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | In vitro methylation of the U7 snRNP subunits Lsm11 and SmE by the PRMT5/MEP50/pICln methylosome.
Rna, 29, 2023
|
|
2OJ9
 
 | | Structure of IGF-1R kinase domain complexed with a benzimidazole inhibitor | | Descriptor: | 3-[5-(1H-IMIDAZOL-1-YL)-7-METHYL-1H-BENZIMIDAZOL-2-YL]-4-[(PYRIDIN-2-YLMETHYL)AMINO]PYRIDIN-2(1H)-ONE, Insulin-like growth factor 1 receptor precursor (EC 2.7.10.1) (Insulin-like growth factor I receptor) (IGF-I receptor) (CD221 antigen) | | Authors: | Sack, J.S, Jacobson, B.L. | | Deposit date: | 2007-01-12 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and initial SAR of 3-(1H-benzo[d]imidazol-2-yl)pyridin-2(1H)-ones as inhibitors of insulin-like growth factor 1-receptor (IGF-1R).
Bioorg.Med.Chem.Lett., 17, 2007
|
|
2RBE
 
 | | The discovery of 2-anilinothiazolones as 11beta-HSD1 inhibitors | | Descriptor: | (5R)-2-[(2-fluorophenyl)amino]-5-(1-methylethyl)-1,3-thiazol-4(5H)-one, Corticosteroid 11-beta-dehydrogenase isozyme 1, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, J, Jordan, S.R, Li, V. | | Deposit date: | 2007-09-18 | | Release date: | 2008-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The discovery of 2-anilinothiazolones as 11beta-HSD1 inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
