2RLI
 
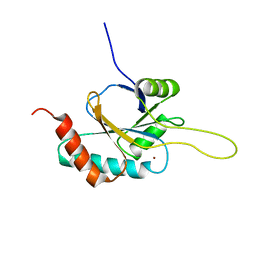 | | Solution structure of Cu(I) human Sco2 | | Descriptor: | COPPER (I) ION, SCO2 protein homolog, mitochondrial | | Authors: | Banci, L, Bertini, I, Ciofi-baffoni, S, Gerothanassis, I.P, Leontari, I, Martinelli, M, Wang, S, Structural Proteomics in Europe (SPINE), Structural Proteomics in Europe 2 (SPINE-2) | | Deposit date: | 2007-07-11 | | Release date: | 2007-08-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A Structural Characterization of Human SCO2
Structure, 15, 2007
|
|
2RHH
 
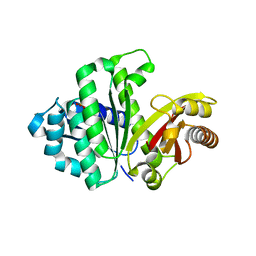 | | Synthetic Gene Encoded Bacillus Subtilis FtsZ with Bound Sulfate Ion | | Descriptor: | Cell Division Protein ftsZ, SULFATE ION | | Authors: | Lovell, S, Halloran, Z, Hjerrild, K, Sheridan, D, Burgin, A, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-10-09 | | Release date: | 2008-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Combined protein construct and synthetic gene engineering for heterologous protein expression and crystallization using Gene Composer.
BMC Biotechnol., 9, 2009
|
|
2RNW
 
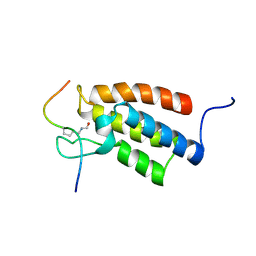 | | The Structural Basis for Site-Specific Lysine-Acetylated Histone Recognition by the Bromodomains of the Human Transcriptional Co-Activators PCAf and CBP | | Descriptor: | Histone H3, Histone acetyltransferase PCAF | | Authors: | Zeng, L, Zhang, Q, Gerona-Navarro, G, Zhou, M.M. | | Deposit date: | 2008-02-03 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Site-Specific Histone Recognition by the Bromodomains of Human Coactivators PCAF and CBP/p300
Structure, 16, 2008
|
|
2RHL
 
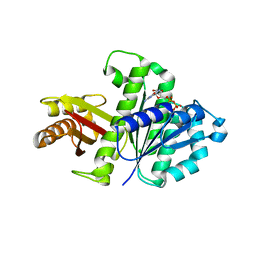 | | Synthetic Gene Encoded Bacillus Subtilis FtsZ NCS Dimer with Bound GDP | | Descriptor: | Cell Division Protein ftsZ, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Lovell, S, Halloran, Z, Hjerrild, K, Sheridan, D, Burgin, A, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D) | | Deposit date: | 2007-10-09 | | Release date: | 2008-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Combined protein construct and synthetic gene engineering for heterologous protein expression and crystallization using Gene Composer.
BMC Biotechnol., 9, 2009
|
|
7YUJ
 
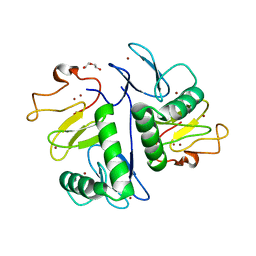 | | Crystal structure of HOIL-1L(365-510) | | Descriptor: | DI(HYDROXYETHYL)ETHER, RanBP-type and C3HC4-type zinc finger-containing protein 1, ZINC ION | | Authors: | Xiao, L, Pan, L. | | Deposit date: | 2022-08-17 | | Release date: | 2023-08-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.865 Å) | | Cite: | Mechanistic insights into the enzymatic activity of E3 ligase HOIL-1L and its regulation by the linear ubiquitin chain binding.
Sci Adv, 9, 2023
|
|
7YUI
 
 | |
2RMI
 
 | | 3D NMR structure of astressin | | Descriptor: | astressin | | Authors: | Royappa, G.C.R, Cervini, L, Gulyas, J, Rivier, J, Riek, R. | | Deposit date: | 2007-10-17 | | Release date: | 2007-10-30 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | Astressin-amide and astressin-acid are structurally different in dimethylsulfoxide
Biopolymers, 87, 2007
|
|
2TSA
 
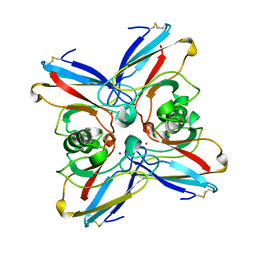 | | AZURIN MUTANT M121A | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Tsai, L.-C, Bonander, N, Harata, K, Karlsson, B.G, Vanngard, T, Langer, V, Sjolin, L. | | Deposit date: | 1996-05-10 | | Release date: | 1996-11-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mutant Met121Ala of Pseudomonas aeruginosa azurin and its azide derivative: crystal structures and spectral properties.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2RD3
 
 | |
2TSB
 
 | | AZURIN MUTANT M121A-AZIDE | | Descriptor: | AZIDE ION, AZURIN AZIDE, COPPER (II) ION | | Authors: | Tsai, L.-C, Bonander, N, Harata, K, Karlsson, B.G, Vanngard, T, Langer, V, Sjolin, L. | | Deposit date: | 1996-05-10 | | Release date: | 1996-11-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mutant Met121Ala of Pseudomonas aeruginosa azurin and its azide derivative: crystal structures and spectral properties.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2RDA
 
 | |
2RHK
 
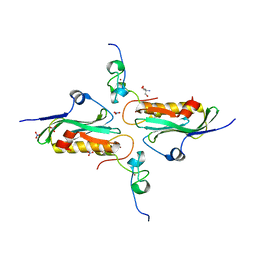 | | Crystal structure of influenza A NS1A protein in complex with F2F3 fragment of human cellular factor CPSF30, Northeast Structural Genomics Targets OR8C and HR6309A | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cleavage and polyadenylation specificity factor subunit 4, NITRATE ION, ... | | Authors: | Das, K, Ma, L.-C, Xiao, R, Radvansky, B, Aramini, J, Zhao, L, Arnold, E, Krug, R.M, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-10-09 | | Release date: | 2008-07-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for suppression of a host antiviral response by influenza A virus.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2RNX
 
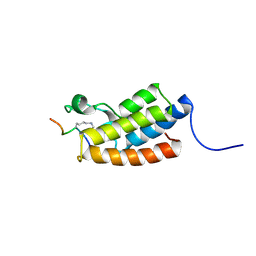 | | The Structural Basis for Site-Specific Lysine-Acetylated Histone Recognition by the Bromodomains of the HUman Transcriptional Co-Activators PCAF and CBP | | Descriptor: | Histone H3, Histone acetyltransferase PCAF | | Authors: | Zeng, L, Zhang, Q, Gerona-Navarro, G, Zhou, M.M. | | Deposit date: | 2008-02-03 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of Site-Specific Histone Recognition by the Bromodomains of Human Coactivators PCAF and CBP/p300
Structure, 16, 2008
|
|
2RJH
 
 | | Crystal structure of biosynthetic alaine racemase in D-cycloserine-bound form from Escherichia coli | | Descriptor: | Alanine racemase, D-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL]-N,O-CYCLOSERYLAMIDE, SULFATE ION | | Authors: | Wu, D, Hu, T, Zhang, L, Jiang, H, Shen, X. | | Deposit date: | 2007-10-15 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Residues Asp164 and Glu165 at the substrate entryway function potently in substrate orientation of alanine racemase from E. coli: Enzymatic characterization with crystal structure analysis
Protein Sci., 17, 2008
|
|
2RQS
 
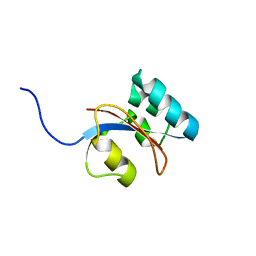 | | 3D structure of Pin from the psychrophilic archeon Cenarcheaum symbiosum (CsPin) | | Descriptor: | Parvulin-like peptidyl-prolyl isomerase | | Authors: | Zhukov, I, Jaremko, L, Jaremko, M, Mueller, J.W, Bayer, P. | | Deposit date: | 2009-11-17 | | Release date: | 2010-11-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure and Dynamics of the First Archaeal Parvulin Reveal a New Functionally Important Loop in Parvulin-type Prolyl Isomerases
J.Biol.Chem., 286, 2011
|
|
7ZPG
 
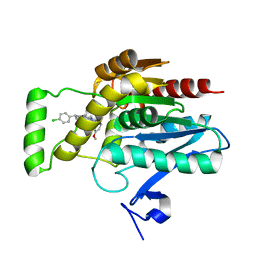 | | CRYSTAL STRUCTURE OF HUMAN MONOGLYCERIDE LIPASE WITH LIGAND | | Descriptor: | Monoglyceride lipase, [(7R,9aR)-7-(4-chlorophenyl)-1,3,4,6,7,8,9,9a-octahydropyrido[1,2-a]pyrazin-2-yl]-(2-bromanyl-3-methoxy-phenyl)methanone | | Authors: | Kemble, A, Hornsperger, B, Ruf, I, Richter, H, Benz, J, Kuhn, B, Heer, D, Wittwer, M, Engelhardt, B, Grether, U, Collin, L, Leibrock, L. | | Deposit date: | 2022-04-27 | | Release date: | 2022-09-21 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | A potent and selective inhibitor for the modulation of MAGL activity in the neurovasculature.
Plos One, 17, 2022
|
|
4V9S
 
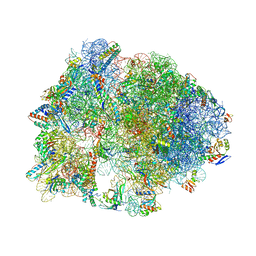 | | Crystal structure of antibiotic GE82832 bound to 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S Ribosomal Protein S10, ... | | Authors: | Bulkley, D.P, Brandi, L, Polikanov, Y.S, Fabbretti, A, O'Connor, M, Gualerzi, C.O, Steitz, T.A. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-09 | | Last modified: | 2018-06-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The antibiotics dityromycin and GE82832 bind protein S12 and block EF-G-catalyzed translocation.
Cell Rep, 6, 2014
|
|
4V7R
 
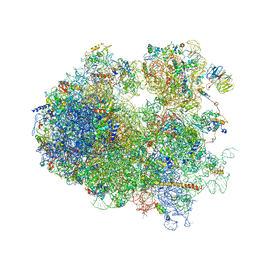 | | Yeast 80S ribosome. | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Ben-Shem, A, Jenner, L, Yusupova, G, Yusupov, M. | | Deposit date: | 2010-07-23 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Crystal structure of the eukaryotic ribosome.
Science, 330, 2010
|
|
4P3Y
 
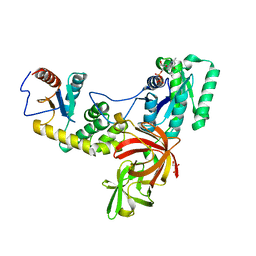 | |
4WBX
 
 | | Conserved hypothetical protein PF1771 from Pyrococcus furiosus solved by sulfur SAD using Swiss Light Source data | | Descriptor: | 2-keto acid:ferredoxin oxidoreductase subunit alpha | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-09-04 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4PIV
 
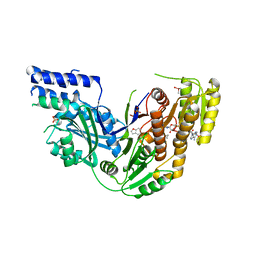 | | Human Fatty Acid Synthase Psi/KR Tri-Domain with NADPH and GSK2194069 | | Descriptor: | 4-[4-(1-benzofuran-5-yl)phenyl]-5-{[(3S)-1-(cyclopropylcarbonyl)pyrrolidin-3-yl]methyl}-2,4-dihydro-3H-1,2,4-triazol-3-one, CACODYLATE ION, Fatty acid synthase, ... | | Authors: | Williams, S.P, Wang, L, Brown, K.K, Parrish, C.A, Hardwicke, M.A. | | Deposit date: | 2014-05-09 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | A human fatty acid synthase inhibitor binds beta-ketoacyl reductase in the keto-substrate site.
Nat.Chem.Biol., 10, 2014
|
|
4UXB
 
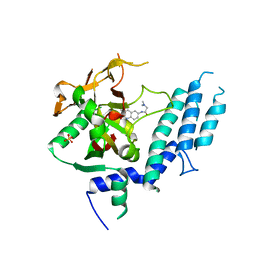 | | Human ARTD1 (PARP1) - Catalytic domain in complex with inhibitor PJ34 | | Descriptor: | N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, POLY ADP-RIBOSE POLYMERASE 1, SULFATE ION | | Authors: | Tresaugues, L, Thorsell, A.G, Karlberg, T, Schuler, H. | | Deposit date: | 2014-08-21 | | Release date: | 2015-09-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Structural Basis for Potency and Promiscuity in Poly(ADP-ribose) Polymerase (PARP) and Tankyrase Inhibitors.
J. Med. Chem., 60, 2017
|
|
4V9C
 
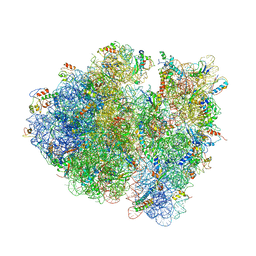 | | Allosteric control of the ribosome by small-molecule antibiotics | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Cate, J.H.D, Pulk, A, Blanchard, S.C, Wang, L, Feldman, M.B, Wasserman, M.R, Altman, R. | | Deposit date: | 2012-07-25 | | Release date: | 2014-07-09 | | Last modified: | 2014-12-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Allosteric control of the ribosome by small-molecule antibiotics.
Nat.Struct.Mol.Biol., 19, 2012
|
|
4V3E
 
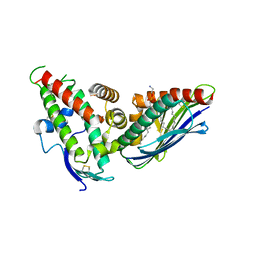 | | The CIDRa domain from IT4var07 PfEMP1 bound to endothelial protein C receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ENDOTHELIAL PROTEIN C RECEPTOR, ... | | Authors: | Lau, C.K.Y, Turner, L, Jespersen, J.S, Lowe, E.D, Petersen, B, Wang, C.W, Petersen, J.E.V, Lusingu, J, Theander, T.G, Lavstsen, T, Higgins, M.K. | | Deposit date: | 2014-10-17 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Conservation Despite Huge Sequence Diversity Allows Epcr Binding by the Pfemp1 Family Implicated in Severe Childhood Malaria.
Cell Host Microbe., 17, 2015
|
|
4WB3
 
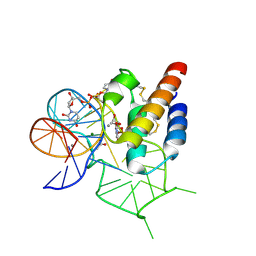 | | Crystal structure of the mirror-image L-RNA/L-DNA aptamer NOX-D20 in complex with mouse C5a-desArg complement anaphylatoxin | | Descriptor: | ACETATE ION, CALCIUM ION, Complement C5, ... | | Authors: | Yatime, L, Maasch, C, Hoehlig, K, Klussmann, S, Vater, A, Andersen, G.R. | | Deposit date: | 2014-09-02 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the targeting of complement anaphylatoxin C5a using a mixed L-RNA/L-DNA aptamer.
Nat Commun, 6, 2015
|
|
