7ZLY
 
 | | Crystal structure of human GPCR Niacin receptor (HCA2) expressed from Spodoptera frugiperda | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Hydroxycarboxylic acid receptor 2,Soluble cytochrome b562, OLEIC ACID | | Authors: | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, FiBerto, J.F, Wu, L.J, Tong, J.H, Han, G.W, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | Deposit date: | 2022-04-17 | | Release date: | 2023-04-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
7XK2
 
 | | Cryo-EM Structure of Human Niacin Receptor HCA2-Gi protein complex | | Descriptor: | 2-[[2,2-dimethyl-3-[3-(5-oxidanylpyridin-2-yl)-1,2,4-oxadiazol-5-yl]propanoyl]amino]cyclohexene-1-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, Y, Kang, H.J, Gao, R.G, Wang, J.J, Han, G.W, DiBerto, J.F, Wu, L.J, Tong, J.H, Qu, L, Wu, Y.R, Pileski, R, Li, X.M, Zhang, X.C, Zhao, S.W, Kenakin, T, Wang, Q, Stevens, R.C, Peng, W, Roth, B.L, Rao, Z.H, Liu, Z.J. | | Deposit date: | 2022-04-19 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the human niacin receptor HCA2-G i signalling complex.
Nat Commun, 14, 2023
|
|
7YF5
 
 | |
7Y3O
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with human antibody BIOLS56 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of BIOLS56, Light chain of BIOLS56, ... | | Authors: | Rao, X, Gao, F, Wu, Y, Gao, F. | | Deposit date: | 2022-06-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7Y3N
 
 | | Crystal structure of SARS-CoV receptor binding domain in complex with human antibody BIOLS56 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of BIOLS56, ... | | Authors: | Rao, X, Chai, Y, Wu, Y, Gao, F. | | Deposit date: | 2022-06-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HRD
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 Delta variant in complex with IMCAS74 Fab and W14 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, IMCAS74 Fab heavy chain, IMCAS74 Fab light chain, ... | | Authors: | Zhao, R.C, Wu, L.L, Han, P. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Defining a de novo non-RBM antibody as RBD-8 and its synergistic rescue of immune-evaded antibodies to neutralize Omicron SARS-CoV-2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5H2D
 
 | | Crystal structure of Osh1 ORD domain in complex with ergosterol | | Descriptor: | ERGOSTEROL, KLLA0C04147p, SULFATE ION | | Authors: | Im, Y.J, Manik, M.K, Yang, H.S, Tong, J.S. | | Deposit date: | 2016-10-14 | | Release date: | 2017-05-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Yeast OSBP-Related Protein Osh1 Reveals Key Determinants for Lipid Transport and Protein Targeting at the Nucleus-Vacuole Junction
Structure, 25, 2017
|
|
5H2A
 
 | |
5H28
 
 | |
5H2C
 
 | | Crystal structure of Saccharomyces cerevisiae Osh1 ANK - Nvj1 | | Descriptor: | Nucleus-vacuole junction protein 1, Oxysterol-binding protein homolog 1 | | Authors: | Im, Y.J, Manik, M.K, Yang, H.S, Tong, J.S. | | Deposit date: | 2016-10-14 | | Release date: | 2017-05-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.508 Å) | | Cite: | Structure of Yeast OSBP-Related Protein Osh1 Reveals Key Determinants for Lipid Transport and Protein Targeting at the Nucleus-Vacuole Junction
Structure, 25, 2017
|
|
5WVR
 
 | | Crystal structure of Osh1 ORD domain in complex with cholesterol | | Descriptor: | CHOLESTEROL, KLLA0C04147p, SULFATE ION | | Authors: | Im, Y.J, Manik, M.K, Yang, H.S, Tong, J.S. | | Deposit date: | 2016-12-28 | | Release date: | 2017-05-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Yeast OSBP-Related Protein Osh1 Reveals Key Determinants for Lipid Transport and Protein Targeting at the Nucleus-Vacuole Junction
Structure, 25, 2017
|
|
6ML1
 
 | | Structure of the USP15 deubiquitinase domain in complex with an affinity-matured inhibitory Ubv | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Singer, A.U, Teyra, J, Boehmelt, G, Lenter, M, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2018-09-26 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Characterization of Ubiquitin Variant Inhibitors of USP15.
Structure, 27, 2019
|
|
7T1K
 
 | | Crystal structure of a superbinder Fes SH2 domain (sFes1) in complex with a high affinity phosphopeptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MALONATE ION, ... | | Authors: | Martyn, G.D, Singer, A.U, Veggiani, G, Kurinov, I, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-12-02 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Engineered SH2 Domains for Targeted Phosphoproteomics.
Acs Chem.Biol., 17, 2022
|
|
7T1L
 
 | | Crystal structure of a superbinder Fes SH2 domain (sFesS) in complex with a high affinity phosphopeptide | | Descriptor: | CHLORIDE ION, SODIUM ION, Synthetic phosphotyrosine-containing Ezrin-derived peptide, ... | | Authors: | Martyn, G.D, Singer, A.U, Veggiani, G, Kurinov, I, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-12-02 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Engineered SH2 Domains for Targeted Phosphoproteomics.
Acs Chem.Biol., 17, 2022
|
|
7T1U
 
 | | Crystal structure of a superbinder Src SH2 domain (sSrcF) in complex with a high affinity phosphopeptide | | Descriptor: | Proto-oncogene tyrosine-protein kinase Src, Synthetic phosphopeptide, ZINC ION | | Authors: | Martyn, G.D, Singer, A.U, Manczyk, N, Veggiani, G, Kurinov, I, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-12-02 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Engineered SH2 Domains for Targeted Phosphoproteomics.
Acs Chem.Biol., 17, 2022
|
|
6CPM
 
 | | Structure of the USP15 deubiquitinase domain in complex with a third-generation inhibitory Ubv | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Singer, A.U, Teyra, J, Boehmelt, G, Lenter, M, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2018-03-13 | | Release date: | 2019-01-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.011 Å) | | Cite: | Structural and Functional Characterization of Ubiquitin Variant Inhibitors of USP15.
Structure, 27, 2019
|
|
6CRN
 
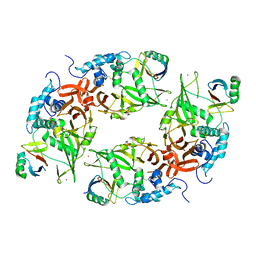 | | Structure of the USP15 deubiquitinase domain in complex with a high-affinity first-generation Ubv | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 15, Ubiquitin variant 15.2, ZINC ION | | Authors: | Singer, A.U, Teyra, J, Boehmelt, G, Lenter, M, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2018-03-19 | | Release date: | 2019-01-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Characterization of Ubiquitin Variant Inhibitors of USP15.
Structure, 27, 2019
|
|
3V6E
 
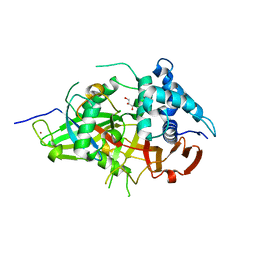 | | Crystal Structure of USP2 and a mutant form of Ubiquitin | | Descriptor: | CHLORIDE ION, GLYCEROL, Ubiquitin, ... | | Authors: | Neculai, M, Ernst, A, Sidhu, S, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-12-19 | | Release date: | 2012-12-19 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A strategy for modulation of enzymes in the ubiquitin system.
Science, 339, 2013
|
|
6DJ9
 
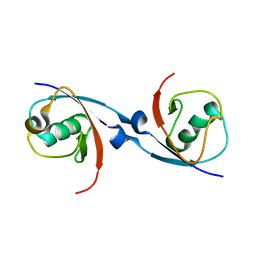 | | Structure of the USP15 DUSP domain in complex with a high-affinity Ubiquitin Variant (UbV) | | Descriptor: | Ubiquitin Variant UbV 15.D, Ubiquitin carboxyl-terminal hydrolase 15 | | Authors: | Singer, A.U, Teyra, J, Boehmelt, G, Lenter, M, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2018-05-24 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and Functional Characterization of Ubiquitin Variant Inhibitors of USP15.
Structure, 27, 2019
|
|
4TZI
 
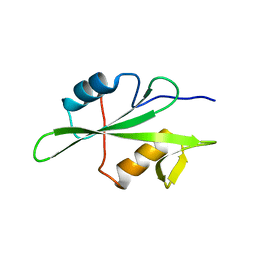 | | Structure of unliganded Lyn SH2 domain | | Descriptor: | Tyrosine-protein kinase Lyn | | Authors: | Wybenga-Groot, L.E. | | Deposit date: | 2014-07-10 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Tyrosine Phosphorylation of the Lyn Src Homology 2 (SH2) Domain Modulates Its Binding Affinity and Specificity.
Mol.Cell Proteomics, 14, 2015
|
|
1JPA
 
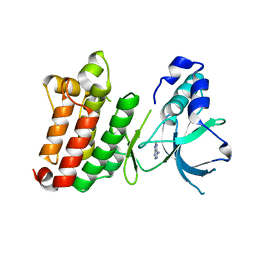 | | Crystal Structure of unphosphorylated EphB2 receptor tyrosine kinase and juxtamembrane region | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, neural kinase, Nuk=Eph/Elk/Eck family receptor-like tyrosine kinase | | Authors: | Wybenga-Groot, L.E, Pawson, T, Sicheri, F. | | Deposit date: | 2001-08-01 | | Release date: | 2001-10-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural basis for autoinhibition of the Ephb2 receptor tyrosine kinase by the unphosphorylated juxtamembrane region.
Cell(Cambridge,Mass.), 106, 2001
|
|
4N9N
 
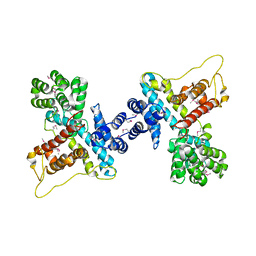 | |
4I6L
 
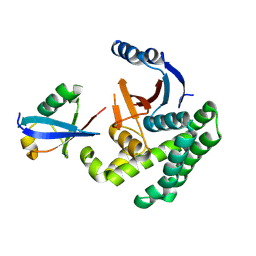 | |
4J7P
 
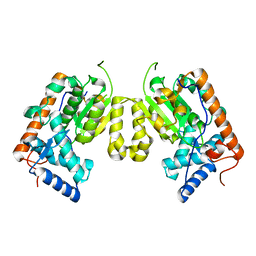 | | Crystal structure of Saccharomyces cerevisiae Sfh3 | | Descriptor: | Phosphatidylinositol transfer protein PDR16 | | Authors: | Yang, H, Im, Y.J. | | Deposit date: | 2013-02-14 | | Release date: | 2013-07-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural determinants for phosphatidylinositol recognition by Sfh3 and substrate-induced dimer-monomer transition during lipid transfer cycles.
Febs Lett., 587, 2013
|
|
4J7Q
 
 | | Crystal structure of Saccharomyces cerevisiae Sfh3 complexed with phosphatidylinositol | | Descriptor: | (1R)-2-{[(S)-hydroxy{[(1S,2R,3R,4S,5S,6R)-2,3,4,5,6-pentahydroxycyclohexyl]oxy}phosphoryl]oxy}-1-[(octadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Phosphatidylinositol transfer protein PDR16 | | Authors: | Yang, H, Im, Y.J. | | Deposit date: | 2013-02-14 | | Release date: | 2013-07-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural determinants for phosphatidylinositol recognition by Sfh3 and substrate-induced dimer-monomer transition during lipid transfer cycles.
Febs Lett., 587, 2013
|
|
