1DB2
 
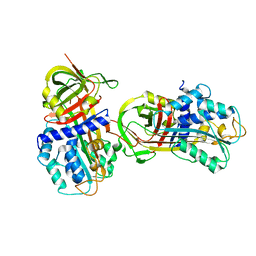 | | CRYSTAL STRUCTURE OF NATIVE PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Descriptor: | PLASMINOGEN ACTIVATOR INHIBITOR-1 | | Authors: | Nar, H, Bauer, M, Stassen, J.M, Lang, D, Gils, A, Declerck, P. | | Deposit date: | 1999-11-02 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Plasminogen activator inhibitor 1. Structure of the native serpin, comparison to its other conformers and implications for serpin inactivation.
J.Mol.Biol., 297, 2000
|
|
3HI9
 
 | |
1MHS
 
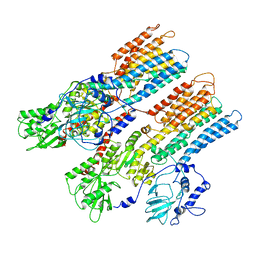 | | Model of Neurospora crassa proton ATPase | | Descriptor: | Plasma Membrane ATPase | | Authors: | Kuhlbrandt, W. | | Deposit date: | 2002-08-21 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | ELECTRON CRYSTALLOGRAPHY (8 Å) | | Cite: | Structure, mechanism and regulation of the Neurospora plasma membrane H+-ATPase
Science, 297, 2002
|
|
4IXD
 
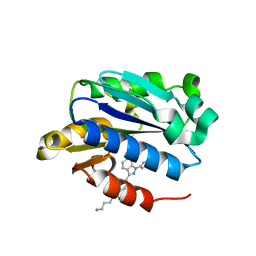 | | X-ray structure of lfa-1 i-domain in complex with ibe-667 at 1.8a resolution | | Descriptor: | 4-(3-{4-[(3-aminopropyl)carbamoyl]phenyl}-1H-indazol-1-yl)-N-methylbenzamide, Integrin alpha-L, MAGNESIUM ION | | Authors: | Kallen, J. | | Deposit date: | 2013-01-25 | | Release date: | 2014-01-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification and x-ray structure based investigation of an ICAM-1 binding enhancing small molecule activator of LFA-1
To be Published, 2013
|
|
4MDK
 
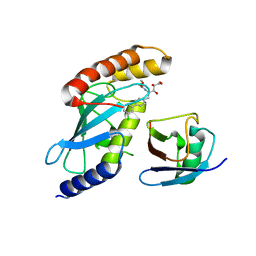 | | Cdc34-ubiquitin-CC0651 complex | | Descriptor: | 4,5-dideoxy-5-(3',5'-dichlorobiphenyl-4-yl)-4-[(methoxyacetyl)amino]-L-arabinonic acid, Ubiquitin, Ubiquitin-conjugating enzyme E2 R1 | | Authors: | Ceccarelli, D.F, Orlicky, S, Tyers, M, Sicheri, F. | | Deposit date: | 2013-08-22 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6095 Å) | | Cite: | E2 enzyme inhibition by stabilization of a low-affinity interface with ubiquitin.
Nat.Chem.Biol., 10, 2014
|
|
1E7P
 
 | | QUINOL:FUMARATE REDUCTASE FROM WOLINELLA SUCCINOGENES | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Lancaster, C.R.D, Kroeger, A. | | Deposit date: | 2000-09-01 | | Release date: | 2001-04-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Third Crystal Form of Wolinella Succinogenes Quinol:Fumarate Reductase Reveals Domain Closure at the Site of Fumarate Reduction
Eur.J.Biochem., 268, 2001
|
|
6FG1
 
 | |
6FG2
 
 | | CRYSTAL STRUCTURE OF FAB OF NATALIZUMAB IN COMPLEX WITH FAB OF NAA84. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, HEAVY CHAIN FAB NAA84, HEAVY CHAIN FAB NATALIZUMAB, ... | | Authors: | Bertrand, T, Pouzieux, S. | | Deposit date: | 2018-01-09 | | Release date: | 2019-07-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.788 Å) | | Cite: | A single T cell epitope drives the neutralizing anti-drug antibody response to natalizumab in multiple sclerosis patients.
Nat. Med., 25, 2019
|
|
2HLE
 
 | |
2BS3
 
 | |
2BS4
 
 | | GLU C180 -> ILE VARIANT QUINOL:FUMARATE REDUCTASE FROMWOLINELLA SUCCINOGENES | | Descriptor: | 2,3-DIMETHYL-1,4-NAPHTHOQUINONE, CITRIC ACID, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Lancaster, C.R.D. | | Deposit date: | 2005-05-14 | | Release date: | 2005-12-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Experimental Support for the E-Pathway Hypothesis of Coupled Transmembrane Electron and Proton Transfer in Dihemic Quinol:Fumarate Reductase
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
6PFK
 
 | |
2BS2
 
 | | QUINOL:FUMARATE REDUCTASE FROM WOLINELLA SUCCINOGENES | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Lancaster, C.R.D. | | Deposit date: | 2005-05-14 | | Release date: | 2006-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Evidence for Transmembrane Proton Transfer in a Dihaem-Containing Membrane Protein Complex.
Embo J., 25, 2006
|
|
6Z35
 
 | | De-novo Maquette 2 protein with buried ion-pair | | Descriptor: | Maquette 2-1ip | | Authors: | Baumgart, M, Roepke, M, Muehlbauer, M, Asami, S, Mader, S, Fredriksson, K, Groll, M, Gamiz-Hernandez, A.P, Kaila, V.R.I. | | Deposit date: | 2020-05-19 | | Release date: | 2021-04-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Design of buried charged networks in artificial proteins.
Nat Commun, 12, 2021
|
|
4OGC
 
 | | Crystal structure of the Type II-C Cas9 enzyme from Actinomyces naeslundii | | Descriptor: | ACETATE ION, HNH endonuclease domain protein, MAGNESIUM ION, ... | | Authors: | Jiang, F, Ma, E, Lin, S, Doudna, J.A. | | Deposit date: | 2014-01-15 | | Release date: | 2014-02-12 | | Last modified: | 2014-03-26 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of Cas9 endonucleases reveal RNA-mediated conformational activation.
Science, 343, 2014
|
|
4OGE
 
 | | Crystal structure of the Type II-C Cas9 enzyme from Actinomyces naeslundii | | Descriptor: | HNH endonuclease domain protein, MAGNESIUM ION, SPERMIDINE, ... | | Authors: | Jiang, F, Ma, E, Lin, S, Doudna, J.A. | | Deposit date: | 2014-01-15 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Structures of Cas9 endonucleases reveal RNA-mediated conformational activation.
Science, 343, 2014
|
|
6Z80
 
 | | stimulatory human GTP cyclohydrolase I - GFRP complex | | Descriptor: | 8-OXO-GUANOSINE-5'-TRIPHOSPHATE, GTP cyclohydrolase 1, GTP cyclohydrolase 1 feedback regulatory protein, ... | | Authors: | Ebenhoch, R, Nar, H, Vonck, J. | | Deposit date: | 2020-06-02 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | A hybrid approach reveals the allosteric regulation of GTP cyclohydrolase I.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Z87
 
 | | human GTP cyclohydrolase I | | Descriptor: | GTP cyclohydrolase 1, ZINC ION | | Authors: | Ebenhoch, R, Nar, H. | | Deposit date: | 2020-06-02 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.564 Å) | | Cite: | A hybrid approach reveals the allosteric regulation of GTP cyclohydrolase I.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Z85
 
 | | inhibitory human GTP cyclohydrolase I - GFRP complex | | Descriptor: | 7,8-DIHYDROBIOPTERIN, GTP cyclohydrolase 1, GTP cyclohydrolase 1 feedback regulatory protein, ... | | Authors: | Ebenhoch, R, Nar, H, Vonck, J. | | Deposit date: | 2020-06-02 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A hybrid approach reveals the allosteric regulation of GTP cyclohydrolase I.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ADH
 
 | |
5ADH
 
 | |
1AXG
 
 | | CRYSTAL STRUCTURE OF THE VAL203->ALA MUTANT OF LIVER ALCOHOL DEHYDROGENASE COMPLEXED WITH COFACTOR NAD AND INHIBITOR TRIFLUOROETHANOL SOLVED TO 2.5 ANGSTROM RESOLUTION | | Descriptor: | ALCOHOL DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRIFLUOROETHANOL, ... | | Authors: | Colby, T.D, Chin, J.K, Bahnson, B.J, Goldstein, B.M, Klinman, J.P. | | Deposit date: | 1997-10-15 | | Release date: | 1998-04-15 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A link between protein structure and enzyme catalyzed hydrogen tunneling.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1ADB
 
 | | CRYSTALLOGRAPHIC STUDIES OF ISOSTERIC NAD ANALOGUES BOUND TO ALCOHOL DEHYDROGENASE: SPECIFICITY AND SUBSTRATE BINDING IN TWO TERNARY COMPLEXES | | Descriptor: | 5-BETA-D-RIBOFURANOSYLNICOTINAMIDE ADENINE DINUCLEOTIDE, ALCOHOL DEHYDROGENASE, ETHANOL, ... | | Authors: | Li, H, Hallows, W.A, Punzi, J.S, Pankiewicz, K.W, Watanabe, K.A, Goldstein, B.M. | | Deposit date: | 1993-12-13 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic studies of isosteric NAD analogues bound to alcohol dehydrogenase: specificity and substrate binding in two ternary complexes.
Biochemistry, 33, 1994
|
|
1ADC
 
 | | CRYSTALLOGRAPHIC STUDIES OF ISOSTERIC NAD ANALOGUES BOUND TO ALCOHOL DEHYDROGENASE: SPECIFICITY AND SUBSTRATE BINDING IN TWO TERNARY COMPLEXES | | Descriptor: | 5-BETA-D-RIBOFURANOSYLPICOLINAMIDE ADENINE-DINUCLEOTIDE, ALCOHOL DEHYDROGENASE, ETHANOL, ... | | Authors: | Li, H, Hallows, W.A, Punzi, J.S, Pankiewicz, K.W, Watanabe, K.A, Goldstein, B.M. | | Deposit date: | 1993-12-13 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic studies of isosteric NAD analogues bound to alcohol dehydrogenase: specificity and substrate binding in two ternary complexes.
Biochemistry, 33, 1994
|
|
7ADH
 
 | |
