6XBG
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAW246 | | Descriptor: | 3C-like proteinase, GLYCEROL, SODIUM ION, ... | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-05 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
5X07
 
 | | Crystal structure of FOXA2 DNA binding domain bound to a full consensus DNA site | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*AP*TP*GP*TP*AP*AP*AP*CP*AP*AP*GP*A)-3'), DNA (5'-D(*TP*CP*TP*TP*GP*TP*TP*TP*AP*CP*AP*TP*TP*TP*TP*G)-3'), Hepatocyte nuclear factor 3-beta | | Authors: | Li, J, Guo, M, Zhou, Z, Jiang, L, Chen, X, Qu, L, Wu, D, Chen, Z, Chen, L, Chen, Y. | | Deposit date: | 2017-01-20 | | Release date: | 2017-08-16 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.796 Å) | | Cite: | Structure of the Forkhead Domain of FOXA2 Bound to a Complete DNA Consensus Site
Biochemistry, 56, 2017
|
|
6XA4
 
 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with UAW241 | | Descriptor: | 3C-like proteinase, GLYCEROL, inhibitor UAW241 | | Authors: | Sacco, M, Ma, C, Wang, J, Chen, Y. | | Deposit date: | 2020-06-03 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and inhibition of the SARS-CoV-2 main protease reveal strategy for developing dual inhibitors against M pro and cathepsin L.
Sci Adv, 6, 2020
|
|
8DDI
 
 | |
8DDM
 
 | |
8DGB
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) Q192T Mutant in Complex with Inhibitor GC376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-23 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DCZ
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) M165Y Mutant in Complex with Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DD1
 
 | | SARS-CoV-2 Main Protease (Mpro) H164N Mutant in Complex with Inhibitor GC376 | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Butler, S.G, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DFN
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) H164N Mutant | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Butler, S.G, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-22 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DFE
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144L Mutant | | Descriptor: | 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-22 | | Release date: | 2022-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8DD9
 
 | | Crystal Structure of SARS-CoV-2 Main Protease (Mpro) S144L Mutant in Complex with Inhibitor GC376 | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase nsp5 | | Authors: | Lewandowski, E.M, Jacobs, L.M.C, Hu, Y, Tan, H, Wang, J, Chen, Y. | | Deposit date: | 2022-06-17 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Naturally Occurring Mutations of SARS-CoV-2 Main Protease Confer Drug Resistance to Nirmatrelvir.
Acs Cent.Sci., 9, 2023
|
|
8HYN
 
 | | Bacterial STING from Riemerella anatipestifer | | Descriptor: | CD-NTase-associated protein 12, TETRAETHYLENE GLYCOL | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2023-01-07 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
8HWJ
 
 | | Bacterial STING from Epilithonimonas lactis in complex with 3'3'-c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, CD-NTase-associated protein 12 | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2022-12-30 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
8HWI
 
 | | Bacterial STING from Larkinella arboricola in complex with 3'3'-c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CD-NTase-associated protein 12 | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2022-12-30 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
8HY8
 
 | | Bacterial STING from Epilithonimonas lactis | | Descriptor: | CD-NTase-associated protein 12 | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.568 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
5K9N
 
 | | Structural and Mechanistic Analysis of Drosophila melanogaster Polyamine N acetyltransferase, an enzyme that Catalyzes the Formation of N acetylagmatine | | Descriptor: | Polyamine N acetyltransferase | | Authors: | Dempsey, D.R, Nichols, D.A, Battistini, M.R, Pemberton, O, Ospina, S.R, Zhang, X, Carpenter, A.-M, Chen, Y, Merkler, D.J. | | Deposit date: | 2016-06-01 | | Release date: | 2017-06-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Mechanistic Analysis of Drosophila melanogaster Agmatine N-Acetyltransferase, an Enzyme that Catalyzes the Formation of N-Acetylagmatine.
Sci Rep, 7, 2017
|
|
8HY9
 
 | | Bacterial STING from Riemerella anatipestifer in complex with 3'3'-c-di-GMP | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), CALCIUM ION, CD-NTase-associated protein 12 | | Authors: | Wang, Y.-C, Yang, C.-S, Hou, M.-H, Chen, Y. | | Deposit date: | 2023-01-06 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.462 Å) | | Cite: | Structural insights into the regulation, ligand recognition, and oligomerization of bacterial STING.
Nat Commun, 14, 2023
|
|
5KW2
 
 | | The extra-helical binding site of GPR40 and the structural basis for allosteric agonism and incretin stimulation | | Descriptor: | (3~{S})-3-cyclopropyl-3-[2-[1-[2-[2,2-dimethylpropyl-(6-methylpyridin-2-yl)carbamoyl]-5-methoxy-phenyl]piperidin-4-yl]-1-benzofuran-6-yl]propanoic acid, Free fatty acid receptor 1,Lysozyme,Free fatty acid receptor 1 | | Authors: | Ho, J.D, Chau, B, Rodgers, L, Lu, F, Wilbur, K.L, Otto, K.A, Chen, Y, Song, M, Riley, J.P, Yang, H.-C, Reynolds, N.A, Kahl, S.D, Lewis, A.P, Groshong, C, Madsen, R.E, Conners, K, Linswala, J.P, Gheyi, T, Saflor, M.D, Lee, M.R, Benach, J, Baker, K.A, Montrose-Rafizadeh, C, Genin, M.J, Miller, A.R, Hamdouchi, C. | | Deposit date: | 2016-07-15 | | Release date: | 2018-05-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structural basis for GPR40 allosteric agonism and incretin stimulation.
Nat Commun, 9, 2018
|
|
6VHS
 
 | | Crystal structure of CTX-M-14 in complex with beta-lactamase inhibitor ETX1317 | | Descriptor: | (2R)-({[(3R,6S)-6-carbamoyl-1-formyl-4-methyl-1,2,3,6-tetrahydropyridin-3-yl]amino}oxy)(fluoro)acetic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Sacco, M.D, Chen, Y. | | Deposit date: | 2020-01-10 | | Release date: | 2020-08-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Discovery of an Orally Available Diazabicyclooctane Inhibitor (ETX0282) of Class A, C, and D Serine beta-Lactamases.
J.Med.Chem., 63, 2020
|
|
1Y7H
 
 | | Structural and biochemical studies identify tobacco SABP2 as a methylsalicylate esterase and further implicate it in plant innate immunity, Northeast Structural Genomics Target AR2241 | | Descriptor: | THIOCYANATE ION, salicylic acid-binding protein 2 | | Authors: | Forouhar, F, Yang, Y, Kumar, D, Chen, Y, Fridman, E, Park, S.W, Chiang, Y, Acton, T.B, Montelione, G.T, Pichersky, E, Klessig, D.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-12-08 | | Release date: | 2004-12-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1Y7I
 
 | | Structural and biochemical studies identify tobacco SABP2 as a methylsalicylate esterase and further implicate it in plant innate immunity, Northeast Structural Genomics Target AR2241 | | Descriptor: | 2-HYDROXYBENZOIC ACID, salicylic acid-binding protein 2 | | Authors: | Forouhar, F, Yang, Y, Kumar, D, Chen, Y, Fridman, E, Park, S.W, Chiang, Y, Acton, T.B, Montelione, G.T, Pichersky, E, Klessig, D.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-12-08 | | Release date: | 2004-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
7PKS
 
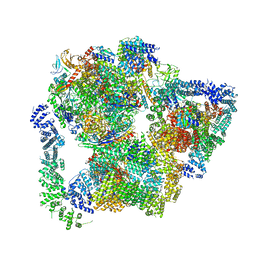 | | Structural basis of Integrator-mediated transcription regulation | | Descriptor: | DNA Template, DNA-directed RNA polymerase II subunit E, DNA-directed RNA polymerase II subunit F, ... | | Authors: | Fianu, I, Chen, Y, Dienemann, C, Cramer, P. | | Deposit date: | 2021-08-26 | | Release date: | 2021-12-08 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of Integrator-mediated transcription regulation.
Science, 374, 2021
|
|
3V3Q
 
 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with Ethyl 2-[2,3,4 trimethoxy-6(1-octanoyl)phenyl]acetate | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1, SODIUM ION, ... | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
7VGI
 
 | |
7VGH
 
 | |
