5Z3U
 
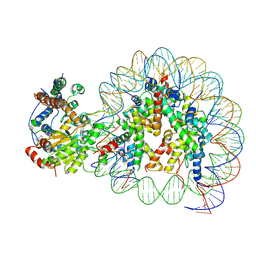 | | Structure of Snf2-nucleosome complex at shl2 in ADP BeFx state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, DNA (167-MER), ... | | Authors: | Li, M, Xia, X, Liu, X, Li, X, Chen, Z. | | Deposit date: | 2018-01-08 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Mechanism of DNA translocation underlying chromatin remodelling by Snf2.
Nature, 567, 2019
|
|
3K3O
 
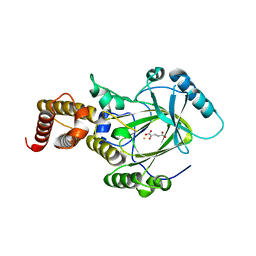 | | Crystal structure of the catalytic core domain of human PHF8 complexed with alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, FE (II) ION, PHD finger protein 8 | | Authors: | Yu, L, Wang, Y, Huang, S, Wang, J, Deng, Z, Wu, W, Gong, W, Chen, Z. | | Deposit date: | 2009-10-03 | | Release date: | 2010-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into a novel histone demethylase PHF8
Cell Res., 20, 2010
|
|
7VWA
 
 | |
7VW8
 
 | |
7VW9
 
 | |
4L4T
 
 | | Structure of human MAIT TCR in complex with human MR1-6-FP | | Descriptor: | 2-amino-4-oxo-3,4-dihydropteridine-6-carbaldehyde, Beta-2-microglobulin, MAIT T-cell receptor alpha chain, ... | | Authors: | Patel, O, Kjer-Nielsen, L, Le Nours, J, Eckle, S.B.G, Birkinshaw, R.W, Beddoe, T, Corbett, A.J, Liu, L, Miles, J.J, Meehan, B, Reantragoon, R, Sandoval-Romero, M.L, Sullivan, L.C, Brooks, A.G, Chen, Z, Fairlie, D.P, McCluskey, J, Rossjohn, J. | | Deposit date: | 2013-06-09 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Recognition of vitamin B metabolites by mucosal-associated invariant T cells.
Nat Commun, 4, 2013
|
|
6A8G
 
 | | The crystal structure of muPAin-1-IG in complex with muPA-SPD at pH8.5 | | Descriptor: | PHOSPHATE ION, Urokinase-type plasminogen activator chain B, muPAin-1-IG | | Authors: | Wang, D, Yang, Y.S, Jiang, L.G, Huang, M.D, Li, J.Y, Andreasen, P.A, Xu, P, Chen, Z. | | Deposit date: | 2018-07-08 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Suppression of Tumor Growth and Metastases by Targeted Intervention in Urokinase Activity with Cyclic Peptides.
J.Med.Chem., 62, 2019
|
|
4L4V
 
 | | Structure of human MAIT TCR in complex with human MR1-RL-6-Me-7-OH | | Descriptor: | 1-deoxy-1-(7-hydroxy-6-methyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Patel, O, Kjer-Nielsen, L, Le Nours, J, Eckle, S.B.G, Birkinshaw, R.W, Beddoe, T, Corbett, A.J, Liu, L, Miles, J.J, Meehan, B, Reantragoon, R, Sandoval-Romero, M.L, Sullivan, L.C, Brooks, A.G, Chen, Z, Fairlie, D.P, McCluskey, J, Rossjohn, J. | | Deposit date: | 2013-06-09 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Recognition of vitamin B metabolites by mucosal-associated invariant T cells.
Nat Commun, 4, 2013
|
|
4DFI
 
 | | Crystal structure of cell adhesion molecule nectin-2/CD112 mutant FAMP | | Descriptor: | Poliovirus receptor-related protein 2 | | Authors: | Liu, J, Qian, X, Chen, Z, Xu, X, Gao, F, Zhang, S, Zhang, R, Qi, J, Gao, G.F, Yan, J. | | Deposit date: | 2012-01-23 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Cell Adhesion Molecule Nectin-2/CD112 and Its Binding to Immune Receptor DNAM-1/CD226
J.Immunol., 188, 2012
|
|
3QQ4
 
 | | Crystal structure of swine major histocompatibility complex class I SLA-1 0401 and identification of 2009 pandemic swine-origin influenza A H1N1 virus cytotoxic T lymphocyte epitope peptides | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, VP35 | | Authors: | Zhang, N, Qi, J, Gao, F, Pan, X, Chen, R, Li, Q, Chen, Z, Li, X, Xia, C, Gao, G.F. | | Deposit date: | 2011-02-15 | | Release date: | 2011-12-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Crystal structure of swine major histocompatibility complex class I SLA-1 0401 and identification of 2009 pandemic swine-origin influenza A H1N1 virus cytotoxic T lymphocyte epitope peptides.
J.Virol., 85, 2011
|
|
3S7H
 
 | | Structure of thrombin mutant Y225P in the E* form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Prothrombin | | Authors: | Niu, W, Chen, Z, Gandhi, P, Vogt, A, Pozzi, N, Pele, L.A, Zapata, F, Di Cera, E. | | Deposit date: | 2011-05-26 | | Release date: | 2011-07-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic and Kinetic Evidence of Allostery in a Trypsin-like Protease.
Biochemistry, 50, 2011
|
|
3EE0
 
 | | Crystal Structure of the W215A/E217A Mutant of Human Thrombin (space group P2(1)2(1)2(1)) | | Descriptor: | Thrombin heavy chain, Thrombin light chain | | Authors: | Gandhi, P.S, Page, M.J, Chen, Z, Bush-Pelc, L, Di Cera, E. | | Deposit date: | 2008-09-03 | | Release date: | 2009-07-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Molecular Basis for the Kinetic Differences of the W215A/E217A Mutant of Human and Murine Thrombin
To be Published
|
|
4LDJ
 
 | | Crystal Structure of a GDP-bound G12C Oncogenic Mutant of Human GTPase KRas | | Descriptor: | GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Hunter, J.C, Gurbani, D, Chen, Z, Westover, K.D. | | Deposit date: | 2013-06-24 | | Release date: | 2014-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | In situ selectivity profiling and crystal structure of SML-8-73-1, an active site inhibitor of oncogenic K-Ras G12C.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4CQM
 
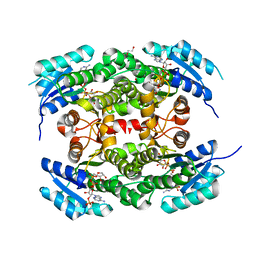 | | Crystal structure of heterotetrameric human ketoacyl reductase complexed with NAD and NADP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CARBONYL REDUCTASE FAMILY MEMBER 4, ... | | Authors: | Venkatesan, R, SahTeli, S.K, Awoniyi, L.O, Jiang, G, Prus, P, Kastoniotis, A.J, Hiltunen, J.K, Wierenga, R.K, Chen, Z. | | Deposit date: | 2014-02-19 | | Release date: | 2014-09-10 | | Last modified: | 2014-09-17 | | Method: | X-RAY DIFFRACTION (2.339 Å) | | Cite: | Insights Into Mitochondrial Fatty Acid Synthesis from the Structure of Heterotetrameric 3-Ketoacyl-Acp Reductase/3R-Hydroxyacyl-Coa Dehydrogenase.
Nat.Commun., 5, 2014
|
|
4DFH
 
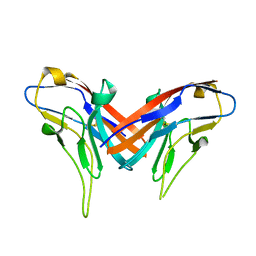 | | Crystal structure of cell adhesion molecule nectin-2/CD112 variable domain | | Descriptor: | Poliovirus receptor-related protein 2 | | Authors: | Liu, J, Qian, X, Chen, Z, Xu, X, Gao, F, Zhang, S, Zhang, R, Qi, J, Gao, G.F, Yan, J. | | Deposit date: | 2012-01-23 | | Release date: | 2012-06-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Cell Adhesion Molecule Nectin-2/CD112 and Its Binding to Immune Receptor DNAM-1/CD226
J.Immunol., 188, 2012
|
|
1JXP
 
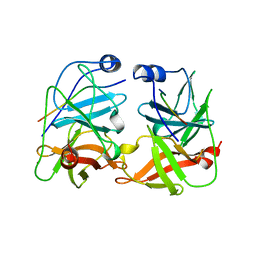 | | BK STRAIN HEPATITIS C VIRUS (HCV) NS3-NS4A | | Descriptor: | NS3 SERINE PROTEASE, NS4A, ZINC ION | | Authors: | Yan, Y, Munshi, S, Chen, Z. | | Deposit date: | 1997-08-21 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Complex of NS3 protease and NS4A peptide of BK strain hepatitis C virus: a 2.2 A resolution structure in a hexagonal crystal form.
Protein Sci., 7, 1998
|
|
3JZ1
 
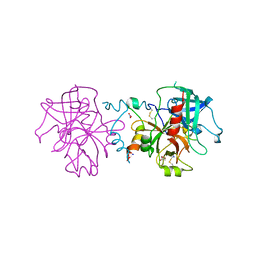 | | Crystal structure of human thrombin mutant N143P in E:Na+ form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, NITRATE ION, ... | | Authors: | Niu, W, Chen, Z, Bush-Pelc, L.A, Bah, A, Gandhi, P.S, Di Cera, E. | | Deposit date: | 2009-09-22 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Mutant N143P reveals how Na+ activates thrombin
J.Biol.Chem., 284, 2009
|
|
3JZ2
 
 | | Crystal structure of human thrombin mutant N143P in E* form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Thrombin heavy chain, ... | | Authors: | Niu, W, Chen, Z, Bush-Pelc, L.A, Bah, A, Gandhi, P.S, Di Cera, E. | | Deposit date: | 2009-09-22 | | Release date: | 2009-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mutant N143P reveals how Na+ activates thrombin
J.Biol.Chem., 284, 2009
|
|
3QDZ
 
 | | Crystal structure of the human thrombin mutant D102N in complex with the extracellular fragment of human PAR4. | | Descriptor: | Proteinase-activated receptor 4, Thrombin heavy chain, Thrombin light chain | | Authors: | Gandhi, P, Chen, Z, Appelbaum, E, Zapata, F, Di Cera, E. | | Deposit date: | 2011-01-19 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of thrombin-protease-receptor interactions
IUBMB LIFE, 63, 2011
|
|
1GG4
 
 | |
1INP
 
 | | CRYSTAL STRUCTURE OF INOSITOL POLYPHOSPHATE 1-PHOSPHATASE AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | INOSITOL POLYPHOSPHATE 1-PHOSPHATASE, MAGNESIUM ION | | Authors: | York, J.D, Ponder, J.W, Chen, Z, Mathews, F.S, Majerus, P.W. | | Deposit date: | 1994-10-04 | | Release date: | 1995-01-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of inositol polyphosphate 1-phosphatase at 2.3-A resolution.
Biochemistry, 33, 1994
|
|
3S7K
 
 | | Structure of thrombin mutant Y225P in the E form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, POTASSIUM ION, Prothrombin | | Authors: | Niu, W, Chen, Z, Gandhi, P, Vogt, A, Pozzi, N, Pele, L.A, Zapata, F, Di Cera, E. | | Deposit date: | 2011-05-26 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic and Kinetic Evidence of Allostery in a Trypsin-like Protease.
Biochemistry, 50, 2011
|
|
3E6P
 
 | | Crystal structure of human meizothrombin desF1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, D-PHENYLALANYL-N-[(1S)-4-{[(Z)-AMINO(IMINO)METHYL]AMINO}-1-(CHLOROACETYL)BUTYL]-L-PROLINAMIDE, Prothrombin, ... | | Authors: | Papaconstantinou, M.E, Gandhi, P, Chen, Z, Bah, A, Di Cera, E. | | Deposit date: | 2008-08-15 | | Release date: | 2008-11-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Na(+) binding to meizothrombin desF1.
Cell.Mol.Life Sci., 65, 2008
|
|
3SJ8
 
 | | Crystal structure of the 3C protease from coxsackievirus A16 | | Descriptor: | 3C protease | | Authors: | Lu, G, Qi, J, Chen, Z, Xu, X, Gao, F, Lin, D, Qian, W, Liu, H, Jiang, H, Yan, J, Gao, G.F. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Enterovirus 71 and Coxsackievirus A16 3C Proteases: Binding to Rupintrivir and Their Substrates and Anti-Hand, Foot, and Mouth Disease Virus Drug Design.
J.Virol., 85, 2011
|
|
3SJO
 
 | | structure of EV71 3C in complex with Rupintrivir (AG7088) | | Descriptor: | 3C protease, 4-{2-(4-FLUORO-BENZYL)-6-METHYL-5-[(5-METHYL-ISOXAZOLE-3-CARBONYL)-AMINO]-4-OXO-HEPTANOYLAMINO}-5-(2-OXO-PYRROLIDIN-3-YL)-PENTANOIC ACID ETHYL ESTER | | Authors: | Lu, G, Qi, J, Chen, Z, Xu, X, Gao, F, Lin, D, Qian, W, Liu, H, Jiang, H, Yan, J, Gao, G.F. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.702 Å) | | Cite: | Enterovirus 71 and Coxsackievirus A16 3C Proteases: Binding to Rupintrivir and Their Substrates and Anti-Hand, Foot, and Mouth Disease Virus Drug Design.
J.Virol., 85, 2011
|
|
