2GST
 
 | | STRUCTURE OF THE XENOBIOTIC SUBSTRATE BINDING SITE OF A GLUTATHIONE S-TRANSFERASE AS REVEALED BY X-RAY CRYSTALLOGRAPHIC ANALYSIS OF PRODUCT COMPLEXES WITH THE DIASTEREOMERS OF 9-(S-GLUTATHIONYL)-10-HYDROXY-9, 10-DIHYDROPHENANTHRENE | | Descriptor: | GLUTATHIONE S-TRANSFERASE, L-gamma-glutamyl-S-[(9S,10S)-10-hydroxy-9,10-dihydrophenanthren-9-yl]-L-cysteinylglycine, SULFATE ION | | Authors: | Ji, X, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1993-06-07 | | Release date: | 1993-10-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and function of the xenobiotic substrate binding site of a glutathione S-transferase as revealed by X-ray crystallographic analysis of product complexes with the diastereomers of 9-(S-glutathionyl)-10-hydroxy-9,10-dihydrophenanthrene.
Biochemistry, 33, 1994
|
|
7MFE
 
 | | Autoinhibited BRAF:(14-3-3)2 complex with the BRAF RBD resolved | | Descriptor: | 14-3-3 protein zeta/delta, Serine/threonine-protein kinase B-raf, ZINC ION | | Authors: | Martinez Fiesco, J.A, Ping, Z, Durrant, D.E, Morrison, D.K. | | Deposit date: | 2021-04-09 | | Release date: | 2022-01-26 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structural insights into the BRAF monomer-to-dimer transition mediated by RAS binding.
Nat Commun, 13, 2022
|
|
7MFD
 
 | | Autoinhibited BRAF:(14-3-3)2:MEK complex with the BRAF RBD resolved | | Descriptor: | 14-3-3 protein zeta/delta, Dual specificity mitogen-activated protein kinase kinase 1, N-(3-fluoro-4-{[4-methyl-2-oxo-7-(pyrimidin-2-yloxy)-2H-chromen-3-yl]methyl}pyridin-2-yl)-N'-methylsulfuric diamide, ... | | Authors: | Martinez Fiesco, J.A, Ping, Z, Durrant, D.E, Morrison, D.K. | | Deposit date: | 2021-04-09 | | Release date: | 2022-01-26 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural insights into the BRAF monomer-to-dimer transition mediated by RAS binding.
Nat Commun, 13, 2022
|
|
7MFF
 
 | | Dimeric (BRAF)2:(14-3-3)2 complex bound to SB590885 Inhibitor | | Descriptor: | (1Z)-5-(2-{4-[2-(DIMETHYLAMINO)ETHOXY]PHENYL}-5-PYRIDIN-4-YL-1H-IMIDAZOL-4-YL)INDAN-1-ONE OXIME, 14-3-3 protein zeta/delta, Serine/threonine-protein kinase B-raf | | Authors: | Martinez Fiesco, J.A, Ping, Z, Durrant, D.E, Morrison, D.K. | | Deposit date: | 2021-04-09 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Structural insights into the BRAF monomer-to-dimer transition mediated by RAS binding.
Nat Commun, 13, 2022
|
|
8WLJ
 
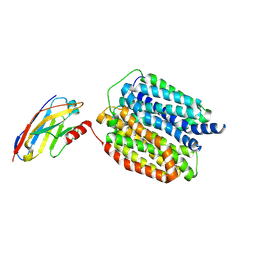 | |
7KNQ
 
 | | SARM1 Octamer | | Descriptor: | NAD(+) hydrolase SARM1 | | Authors: | Shen, C, Wu, H. | | Deposit date: | 2020-11-05 | | Release date: | 2021-11-17 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Multiple domain interfaces mediate SARM1 autoinhibition.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4BJH
 
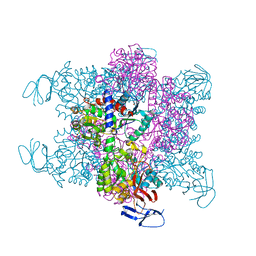 | | Crystal Structure of the Aquifex Reactor Complex Formed by Dihydroorotase (H180A, H232A) with Dihydroorotate and Aspartate Transcarbamoylase with N-(phosphonacetyl)-L-aspartate (PALA) | | Descriptor: | (4S)-2,6-DIOXOHEXAHYDROPYRIMIDINE-4-CARBOXYLIC ACID, 1,2-ETHANEDIOL, ASPARTATE CARBAMOYLTRANSFERASE, ... | | Authors: | Edwards, B.F.P, Martin, P.D, Grimley, E, Vaishnav, A, Fernando, R, Brunzelle, J.S, Cordes, M, Evans, H.G, Evans, D.R. | | Deposit date: | 2013-04-18 | | Release date: | 2013-12-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Mononuclear Metal Center of Type-I Dihydroorotase from Aquifex Aeolicus.
Bmc Biochem., 14, 2013
|
|
8OST
 
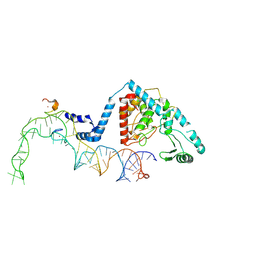 | | Structure of human terminal uridylyltransferase 4 (TUT4, ZCCHC11) in complex with pre-let7g miRNA and Lin28A | | Descriptor: | Protein lin-28 homolog A, Terminal uridylyltransferase 4, ZINC ION, ... | | Authors: | Gilbert, R.J, Yi, G, Ye, M. | | Deposit date: | 2023-04-20 | | Release date: | 2024-07-17 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8OPP
 
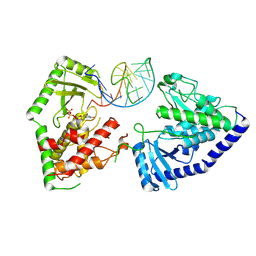 | | Structure of human terminal uridylyltransferase 7 (hTUT7/ZCCHC6) bound with pre-let7g miRNA and UTPalphaS | | Descriptor: | RNA (25-MER), Terminal uridylyltransferase 7, [[(2~{R},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-sulfanyl-phosphoryl] phosphono hydrogen phosphate | | Authors: | Yi, G, Ye, M, Gilbert, R.J. | | Deposit date: | 2023-04-07 | | Release date: | 2024-07-24 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8K19
 
 | |
8OEF
 
 | |
8OPS
 
 | | Human terminal uridylyltransferase 7 (TUT7/ZCCHC6) bound with pre-let7g miRNA and Lin28A - complex 1 | | Descriptor: | Protein lin-28 homolog A, RNA (71-MER) Let7g, Terminal uridylyltransferase 7, ... | | Authors: | Yi, G, Ye, M, Gilbert, R.J. | | Deposit date: | 2023-04-08 | | Release date: | 2024-07-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8OPT
 
 | | Human terminal uridylyltransferase 7 (TUT7/ZCCHC6) bound with pre-let7g miRNA and Lin28A - complex 2 | | Descriptor: | Protein lin-28 homolog A, RNA (53-MER), Terminal uridylyltransferase 7, ... | | Authors: | Yi, G, Ye, M, Gilbert, R.J. | | Deposit date: | 2023-04-08 | | Release date: | 2024-07-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structural basis for activity switching in polymerases determining the fate of let-7 pre-miRNAs.
Nat.Struct.Mol.Biol., 31, 2024
|
|
9ARV
 
 | | CryoEM structure of AMETA-A3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, Isoform 1 of Immunoglobulin heavy constant mu | | Authors: | Huang, W, Sang, Z, Taylor, D. | | Deposit date: | 2024-02-23 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Adaptive multi-epitope targeting and avidity-enhanced nanobody platform for ultrapotent, durable antiviral therapy.
Cell, 187, 2024
|
|
8WLL
 
 | | Cryo-EM structure of human VMAT2 Y422C, in the presence of reserpine, determined in an inward-facing conformation | | Descriptor: | Synaptic vesicular amine transporter, reserpine | | Authors: | Qu, Q, Wang, Y, Zhou, Z. | | Deposit date: | 2023-09-30 | | Release date: | 2024-06-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Transport and inhibition mechanism for VMAT2-mediated synaptic vesicle loading of monoamines.
Cell Res., 34, 2024
|
|
8WLK
 
 | | Cryo-EM structure of human VMAT2 in presence of Tetrabenazine, determined in an outward-facing conformation | | Descriptor: | (3S,5R,11bS)-9,10-dimethoxy-3-(2-methylpropyl)-1,3,4,6,7,11b-hexahydro-2H-pyrido[2,1-a]isoquinolin-2-one, Synaptic vesicular amine transporter | | Authors: | Qu, Q, Wang, Y, Zhou, Z. | | Deposit date: | 2023-09-30 | | Release date: | 2024-06-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Transport and inhibition mechanism for VMAT2-mediated synaptic vesicle loading of monoamines.
Cell Res., 34, 2024
|
|
8WLM
 
 | |
7VEZ
 
 | |
7VF0
 
 | |
7VEY
 
 | | Crystal structure of Cyclosorus parasiticus chalcone synthase 1 (CpCHS1) | | Descriptor: | chalcone synthases | | Authors: | Li, J.X, Cheng, A.X. | | Deposit date: | 2021-09-10 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional and Structural Investigation of Chalcone Synthases Based on Integrated Metabolomics and Transcriptome Analysis on Flavonoids and Anthocyanins Biosynthesis of the Fern Cyclosorus parasiticus .
Front Plant Sci, 12, 2021
|
|
8BCM
 
 | |
7DCR
 
 | | cryo-EM structure of the DEAH-box helicase Prp2 in complex with its coactivator Spp2 | | Descriptor: | PRP2 isoform 1, Pre-mRNA-splicing factor SPP2 | | Authors: | Bai, R, Wan, R, Yan, C, Jia, Q, Zhang, P, Lei, J, Shi, Y. | | Deposit date: | 2020-10-26 | | Release date: | 2021-01-06 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Mechanism of spliceosome remodeling by the ATPase/helicase Prp2 and its coactivator Spp2.
Science, 371, 2021
|
|
7DCO
 
 | | Cryo-EM structure of the activated spliceosome (Bact complex) at an atomic resolution of 2.5 angstrom | | Descriptor: | BJ4_G0014900.mRNA.1.CDS.1, BJ4_G0027490.mRNA.1.CDS.1, BJ4_G0037700.mRNA.1.CDS.1, ... | | Authors: | Bai, R, Wan, R, Yan, C, Qi, J, Zhang, P, Lei, J, Shi, Y. | | Deposit date: | 2020-10-26 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Mechanism of spliceosome remodeling by the ATPase/helicase Prp2 and its coactivator Spp2.
Science, 371, 2021
|
|
7DCQ
 
 | | cryo-EM structure of the DEAH-box helicase Prp2 | | Descriptor: | PRP2 isoform 1 | | Authors: | Bai, R, Wan, R, Yan, C, Jia, Q, Zhang, P, Lei, J, Shi, Y. | | Deposit date: | 2020-10-26 | | Release date: | 2021-01-06 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Mechanism of spliceosome remodeling by the ATPase/helicase Prp2 and its coactivator Spp2.
Science, 371, 2021
|
|
7DD3
 
 | | Cryo-EM structure of the pre-mRNA-loaded DEAH-box ATPase/helicase Prp2 in complex with Spp2 | | Descriptor: | PRP2 isoform 1, Pre-mRNA-splicing factor SPP2, pre-mRNA | | Authors: | Bai, R, Wan, R, Yan, C, Qi, J, Zhang, P, Lei, J, Shi, Y. | | Deposit date: | 2020-10-27 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of spliceosome remodeling by the ATPase/helicase Prp2 and its coactivator Spp2.
Science, 371, 2021
|
|
