6Q6Z
 
 | |
4D1F
 
 | |
1HKA
 
 | | 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE | | Descriptor: | 6-HYDROXYMETHYL-7,8-DIHYDROPTERIN PYROPHOSPHOKINASE | | Authors: | Xiao, B, Shi, G, Chen, X, Yan, H, Ji, X. | | Deposit date: | 1998-09-29 | | Release date: | 1999-06-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase, a potential target for the development of novel antimicrobial agents.
Structure Fold.Des., 7, 1999
|
|
2JLB
 
 | | Xanthomonas campestris putative OGT (XCC0866), complex with UDP- GlcNAc phosphonate analogue | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, URIDINE-DIPHOSPHATE-METHYLENE-N-ACETYL-GLUCOSAMINE, ... | | Authors: | Schuettelkopf, A.W, Clarke, A.J, van Aalten, D.M.F. | | Deposit date: | 2008-09-07 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights Into Mechanism and Specificity of O-Glcnac Transferase.
Embo J., 27, 2008
|
|
6I8H
 
 | |
6I8G
 
 | |
8PPM
 
 | | IL-12Rb1 neutralizing Fab4, crystal kappa variant | | Descriptor: | Fab4 Light chain, crystal kappa variant, Fab4 heavy chain, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-07-07 | | Release date: | 2024-02-07 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8BI2
 
 | | Syk kinase domain in complex with macrocyclic inhibitor 20a | | Descriptor: | 10,13,23-trimethyl-16-oxa-2,4,8,9,13,19,23,30-octazapentacyclo[19.5.2.1^{3,7}.1^{8,11}.0^{24,28}]triaconta-1(27),3,5,7(30),9,11(29),21,24(28),25-nonaen-20-one, Tyrosine-protein kinase SYK | | Authors: | Read, J.A, Patel, J. | | Deposit date: | 2022-11-01 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.508 Å) | | Cite: | Optimization of a series of novel, potent and selective Macrocyclic SYK inhibitors.
Bioorg.Med.Chem.Lett., 91, 2023
|
|
7ZXK
 
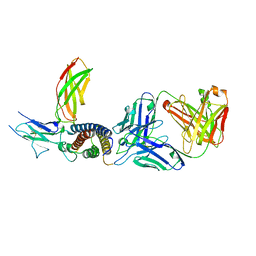 | | Human IL-27 in complex with neutralizing SRF388 FAb fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-27 subunit alpha, Interleukin-27 subunit beta, ... | | Authors: | Bloch, Y, Skladanowska, K, Strand, J, Welin, M, Logan, D, Hill, J, Savvides, S.N. | | Deposit date: | 2022-05-21 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of activation and antagonism of receptor signaling mediated by interleukin-27.
Cell Rep, 41, 2022
|
|
2Q5E
 
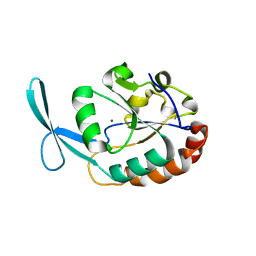 | | Crystal structure of human carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 2 | | Descriptor: | Carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 2, MAGNESIUM ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Lau, C, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-05-31 | | Release date: | 2007-06-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
2R0B
 
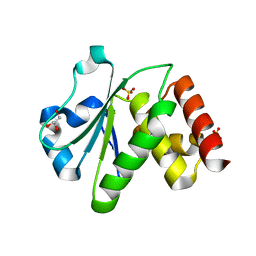 | | Crystal structure of human tyrosine phosphatase-like serine/threonine/tyrosine-interacting protein | | Descriptor: | GLYCEROL, SULFATE ION, Serine/threonine/tyrosine-interacting protein | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Iizuka, M, Romero, R, Smith, D, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-08-18 | | Release date: | 2007-08-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural genomics of protein phosphatases.
J.Struct.Funct.Genom., 8, 2007
|
|
4OQX
 
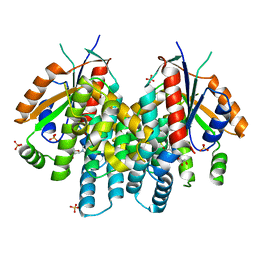 | | Crystal structure of thymidine kinase from herpes simplex virus type 1 in complex with Me-ARA-EdU | | Descriptor: | 1-(2-deoxy-2-methyl-beta-D-arabinofuranosyl)-5-ethynylpyrimidine-2,4(1H,3H)-dione, SULFATE ION, Thymidine kinase | | Authors: | Pernot, L, Neef, A.B, Westermaier, Y, Perozzo, R, Luedtke, N, Scapozza, L. | | Deposit date: | 2014-02-10 | | Release date: | 2014-08-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of HSV1-TK complexed with Me-ARA-EdU
To be Published
|
|
4A5U
 
 | |
3O4N
 
 | |
3O4Q
 
 | |
2HX6
 
 | | Solution structure analysis of the phage T4 endoribonuclease RegB | | Descriptor: | Ribonuclease | | Authors: | Odaert, B, Saida, F, Uzan, M, Bontems, F. | | Deposit date: | 2006-08-02 | | Release date: | 2006-10-31 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and functional studies of RegB, a new member of a family of sequence-specific ribonucleases involved in mRNA inactivation on the ribosome.
J.Biol.Chem., 282, 2007
|
|
2N7D
 
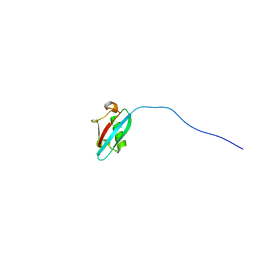 | |
4BPR
 
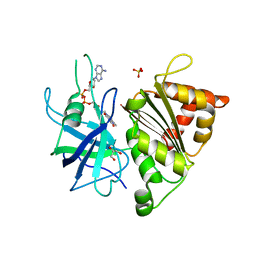 | | FERREDOXIN-NADP REDUCTASE MUTANT WITH TYR 79 REPLACED BY PHE (Y79F) | | Descriptor: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Herguedas, B, Martinez-Julvez, M, Sanchez-Azqueta, A, Hervas, M, Navarro, J.A, Medina, M. | | Deposit date: | 2013-05-28 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Hydrogen Bond Network in the Active Site of Anabaena Ferredoxin-Nadp(+) Reductase Modulates its Catalytic Efficiency.
Biochim.Biophys.Acta, 1837, 2013
|
|
5BOQ
 
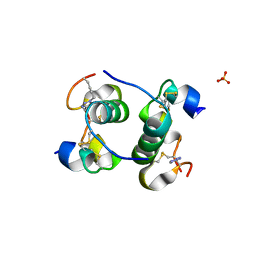 | | Human insulin with intra-chain chemical crosslink between modified B24 and B29 | | Descriptor: | Insulin, SULFATE ION | | Authors: | Brzozowski, A.M, Turkenburg, J.P, Jiracek, J, Zakova, L. | | Deposit date: | 2015-05-27 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational steering of insulin binding specificity by intra-chain chemical crosslinking.
Sci Rep, 6, 2016
|
|
5BPO
 
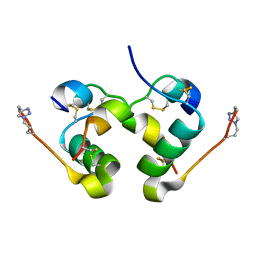 | |
5BQQ
 
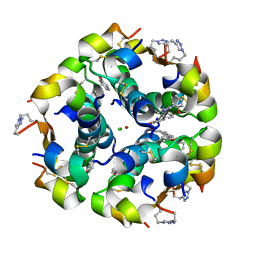 | | Human insulin with intra-chain chemical crosslink between modified B27 and B30 | | Descriptor: | CHLORIDE ION, Insulin, PHENOL, ... | | Authors: | Brzozowski, A.M, Turkenburg, J.P, Jiracek, J, Zakova, L. | | Deposit date: | 2015-05-29 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Rational steering of insulin binding specificity by intra-chain chemical crosslinking.
Sci Rep, 6, 2016
|
|
1VI7
 
 | | Crystal structure of an hypothetical protein | | Descriptor: | Hypothetical protein yigZ | | Authors: | Structural GenomiX | | Deposit date: | 2003-12-01 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of YIGZ, a conserved hypothetical protein from Escherichia coli k12 with a novel fold
Proteins, 55, 2004
|
|
2XWX
 
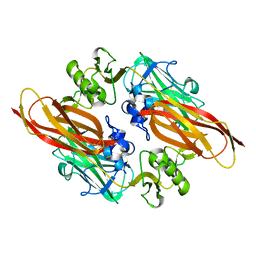 | | Vibrio cholerae colonization factor GbpA crystal structure | | Descriptor: | GLCNAC-BINDING PROTEIN A | | Authors: | Wong, E, Vaaje-Kolstad, G, Ghosh, A, Guerrero, R.H, Konarev, P.V, Ibrahim, A.F.M, Svergun, D.I, Eijsink, V.G.H, Chatterjee, N.S, van Aalten, D.M.F. | | Deposit date: | 2010-11-06 | | Release date: | 2011-11-16 | | Last modified: | 2015-04-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Vibrio Cholerae Colonization Factor Gbpa Possesses a Modular Structure that Governs Binding to Different Host Surfaces.
Plos Pathog., 8, 2012
|
|
7ZG0
 
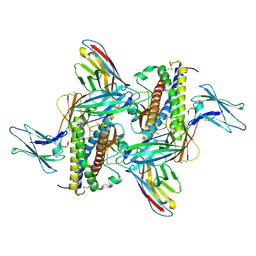 | | Murine IL-27 in complex with IL-27Ra and a non-competing Nb | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-27 receptor subunit alpha, ... | | Authors: | Skladanowska, K, Bloch, Y, Savvides, S.N. | | Deposit date: | 2022-04-01 | | Release date: | 2022-11-02 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Structural basis of activation and antagonism of receptor signaling mediated by interleukin-27.
Cell Rep, 41, 2022
|
|
3Q4F
 
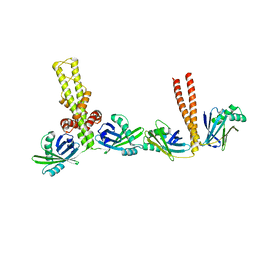 | | Crystal structure of xrcc4/xlf-cernunnos complex | | Descriptor: | DNA repair protein XRCC4, Non-homologous end-joining factor 1 | | Authors: | Ropars, V, Legrand, P, Charbonnier, J.B. | | Deposit date: | 2010-12-23 | | Release date: | 2011-08-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (5.5 Å) | | Cite: | Structural characterization of filaments formed by human Xrcc4-Cernunnos/XLF complex involved in nonhomologous DNA end-joining.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
