3GMN
 
 | | Structure of mouse CD1d in complex with C10Ph | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2 microglobulin, ... | | Authors: | Schiefner, A, Wilson, I.A. | | Deposit date: | 2009-03-14 | | Release date: | 2009-11-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural evaluation of potent NKT cell agonists: implications for design of novel stimulatory ligands.
J.Mol.Biol., 394, 2009
|
|
3GMR
 
 | | Structure of mouse CD1d in complex with C8Ph, different space group | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2 microglobulin, ... | | Authors: | Schiefner, A, Wilson, I.A. | | Deposit date: | 2009-03-14 | | Release date: | 2009-11-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural evaluation of potent NKT cell agonists: implications for design of novel stimulatory ligands.
J.Mol.Biol., 394, 2009
|
|
3GMM
 
 | | Structure of mouse CD1d in complex with C8Ph | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2 microglobulin, ... | | Authors: | Schiefner, A, Wilson, I.A. | | Deposit date: | 2009-03-14 | | Release date: | 2009-11-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural evaluation of potent NKT cell agonists: implications for design of novel stimulatory ligands.
J.Mol.Biol., 394, 2009
|
|
3GZN
 
 | | Structure of NEDD8-activating enzyme in complex with NEDD8 and MLN4924 | | Descriptor: | NEDD8, NEDD8-activating enzyme E1 catalytic subunit, NEDD8-activating enzyme E1 regulatory subunit, ... | | Authors: | Sintchak, M.D. | | Deposit date: | 2009-04-07 | | Release date: | 2010-02-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Substrate-assisted inhibition of ubiquitin-like protein-activating enzymes: the NEDD8 E1 inhibitor MLN4924 forms a NEDD8-AMP mimetic in situ.
Mol.Cell, 37, 2010
|
|
3GMO
 
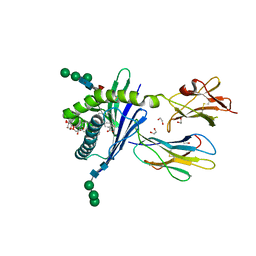 | | Structure of mouse CD1d in complex with C8PhF | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 8-(4-fluorophenyl)-N-{(1S,2S,3R)-1-[(alpha-D-galactopyranosyloxy)methyl]-2,3-dihydroxyheptadecyl}octanamide, ... | | Authors: | Schiefner, A, Wilson, I.A. | | Deposit date: | 2009-03-14 | | Release date: | 2009-11-10 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural evaluation of potent NKT cell agonists: implications for design of novel stimulatory ligands.
J.Mol.Biol., 394, 2009
|
|
5W5D
 
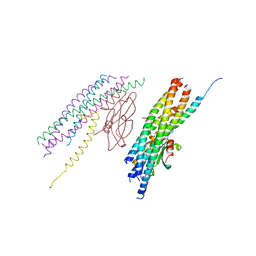 | |
5WTW
 
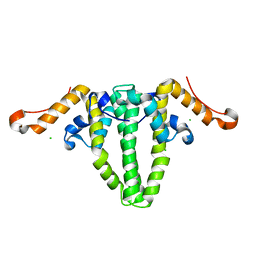 | |
5WQA
 
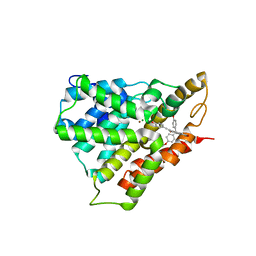 | | Crystal structure of PDE4D catalytic domain complexed with Selaginpulvilins K | | Descriptor: | 1-[2-(4-hydroxyphenyl)ethynyl]-9,9-bis(4-methoxyphenyl)-7-oxidanyl-fluorene-2-carbaldehyde, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huang, Y, Zhang, T, Zheng, X, Yin, S, Luo, H.B. | | Deposit date: | 2016-11-24 | | Release date: | 2017-02-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The discovery, complex crystal structure, and recognition mechanism of a novel natural PDE4 inhibitor from Selaginella pulvinata
Biochem. Pharmacol., 130, 2017
|
|
5UK5
 
 | | Complex of Notch1(EGF8-12) bound to Jagged1(N-EGF3) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Garcia, K.C, Luca, V.C. | | Deposit date: | 2017-01-19 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.506 Å) | | Cite: | Notch-Jagged complex structure implicates a catch bond in tuning ligand sensitivity.
Science, 355, 2017
|
|
5W5C
 
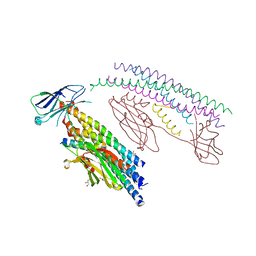 | |
5WRE
 
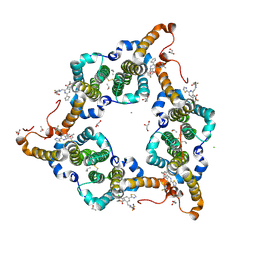 | | Hepatitis B virus core protein Y132A mutant in complex with heteroaryldihydropyrimidine (HAP_R01) | | Descriptor: | (2S)-1-[[(4R)-4-(2-chloranyl-4-fluoranyl-phenyl)-5-methoxycarbonyl-2-(1,3-thiazol-2-yl)-1,4-dihydropyrimidin-6-yl]methyl]-4,4-bis(fluoranyl)pyrrolidine-2-carboxylic acid, CHLORIDE ION, Core protein, ... | | Authors: | Zhou, Z, Xu, Z.H. | | Deposit date: | 2016-12-01 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.946 Å) | | Cite: | Heteroaryldihydropyrimidine (HAP) and Sulfamoylbenzamide (SBA) Inhibit Hepatitis B Virus Replication by Different Molecular Mechanisms.
Sci Rep, 7, 2017
|
|
5ZXD
 
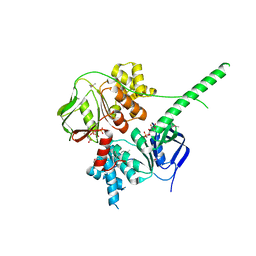 | | Crystal structure of ATP-bound human ABCF1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family F member 1 | | Authors: | Qu, L, Jiang, Y, Liu, Z.J. | | Deposit date: | 2018-05-18 | | Release date: | 2018-07-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Crystal Structure of ATP-Bound Human ABCF1 Demonstrates a Unique Conformation of ABC Proteins
Structure, 26, 2018
|
|
1QNK
 
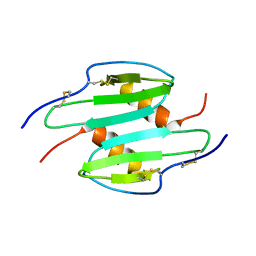 | | TRUNCATED HUMAN GROB[5-73], NMR, 20 STRUCTURES | | Descriptor: | C-X-C motif chemokine 2 | | Authors: | Qian, Y.Q, Johanson, K, McDevitt, P. | | Deposit date: | 1999-10-18 | | Release date: | 2000-02-04 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of truncated human GRObeta [5-73] and its structural comparison with CXC chemokine family members GROalpha and IL-8.
J. Mol. Biol., 294, 1999
|
|
7YAV
 
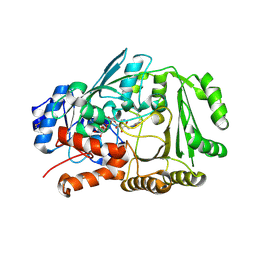 | | Crystal structure of Diels-Alderase MaDA1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, MaDA1 | | Authors: | Lei, X.G, Chen, R.C, Du, X.X, Yang, J, Fan, J.P, Guo, N.X, Ding, Q. | | Deposit date: | 2022-06-28 | | Release date: | 2024-01-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The evolutionary origin of naturally occurring intermolecular Diels-Alderases from Morus alba.
Nat Commun, 15, 2024
|
|
6JMK
 
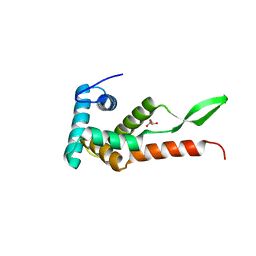 | | Ribosomal protein S7 from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, 30S ribosomal protein S7, GLYCEROL | | Authors: | Li, Z, Li, J. | | Deposit date: | 2019-03-11 | | Release date: | 2019-03-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into the complex of trigger factor chaperone and ribosomal protein S7 from Mycobacterium tuberculosis.
Biochem. Biophys. Res. Commun., 512, 2019
|
|
6I3Y
 
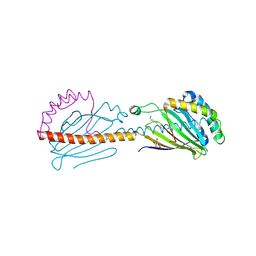 | | Crystal structure of the human mitochondrial PRELID1K58V-TRIAP1 complex with PS | | Descriptor: | DODECYL-BETA-D-MALTOSIDE, O-[(R)-{[(2R)-2,3-bis(octadecanoyloxy)propyl]oxy}(hydroxy)phosphoryl]-L-serine, PRELI domain-containing protein 1, ... | | Authors: | Miliara, X, Berry, J.-L, Morgan, R.M.L, Matthews, S.J. | | Deposit date: | 2018-11-08 | | Release date: | 2019-03-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural determinants of lipid specificity within Ups/PRELI lipid transfer proteins.
Nat Commun, 10, 2019
|
|
2JTK
 
 | |
7YF1
 
 | |
6I4Y
 
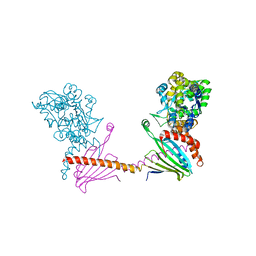 | | X-ray structure of the human mitochondrial PRELID3b-TRIAP1 complex | | Descriptor: | Maltose transport system, substrate-binding protein,TP53-regulated inhibitor of apoptosis 1, PRELI domain containing protein 3B, ... | | Authors: | Miliara, X, Berry, J.-L, Morgan, R.M.L, Matthews, S.J. | | Deposit date: | 2018-11-12 | | Release date: | 2019-03-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural determinants of lipid specificity within Ups/PRELI lipid transfer proteins.
Nat Commun, 10, 2019
|
|
6I3V
 
 | | x-ray structure of the human mitochondrial PRELID1 in complex with TRIAP1 | | Descriptor: | CHLORIDE ION, MYRISTIC ACID, PRELI domain-containing protein 1, ... | | Authors: | Berry, J.L, Miliara, X, Morgan, R.M.L, Matthews, S.J. | | Deposit date: | 2018-11-07 | | Release date: | 2019-03-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural determinants of lipid specificity within Ups/PRELI lipid transfer proteins.
Nat Commun, 10, 2019
|
|
6KI0
 
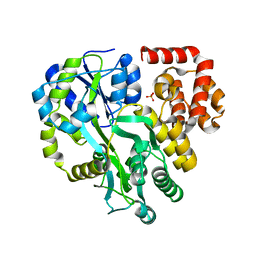 | | Crystal Structure of Human ASC-CARD | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Apoptosis-associated speck-like protein containing a CARD, SULFATE ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Xu, Z.H, Jin, T.C. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Homotypic CARD-CARD interaction is critical for the activation of NLRP1 inflammasome.
Cell Death Dis, 12, 2021
|
|
4IQR
 
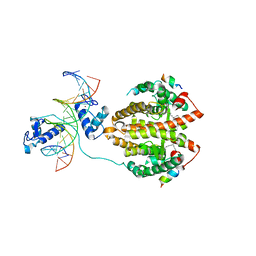 | | Multi-Domain Organization of the HNF4alpha Nuclear Receptor Complex on DNA | | Descriptor: | DNA (5'-D(*CP*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*AP*GP*TP*TP*C)-3'), DNA (5'-D(*GP*GP*AP*AP*CP*TP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*AP*G)-3'), Hepatocyte nuclear factor 4-alpha, ... | | Authors: | Chandra, V, Huang, P, Kim, Y, Rastinejad, F. | | Deposit date: | 2013-01-13 | | Release date: | 2013-03-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Multidomain integration in the structure of the HNF-4 alpha nuclear receptor complex.
Nature, 495, 2013
|
|
7CSD
 
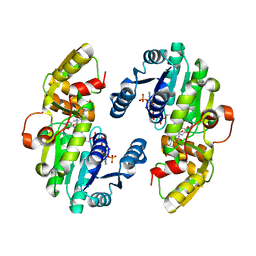 | | AtPrR1 with NADP+ and (+)lariciresinol | | Descriptor: | 4-[[(3R,4R,5S)-4-(hydroxymethyl)-5-(3-methoxy-4-oxidanyl-phenyl)oxolan-3-yl]methyl]-2-methoxy-phenol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol reductase 1 | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79709971 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CS2
 
 | | Apo structure of dimeric IiPLR1 | | Descriptor: | Pinoresinol-lariciresinol reductase | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.68800473 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
7CS6
 
 | | IiPLR1 with NADP+ and (-)lariciresinol | | Descriptor: | 4-[[(3S,4S,5R)-4-(hydroxymethyl)-5-(3-methoxy-4-oxidanyl-phenyl)oxolan-3-yl]methyl]-2-methoxy-phenol, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Pinoresinol-lariciresinol reductase | | Authors: | Shao, K, Zhang, P. | | Deposit date: | 2020-08-14 | | Release date: | 2021-06-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.20139647 Å) | | Cite: | Structure-based engineering of substrate specificity for pinoresinol-lariciresinol reductases.
Nat Commun, 12, 2021
|
|
