8OIZ
 
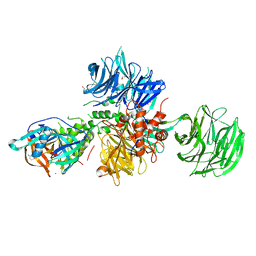 | | Crystal structure of human CRBN-DDB1 in complex with Pomalidomide | | Descriptor: | 1,2-ETHANEDIOL, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Le Bihan, Y.-V, Cabry, M.P, van Montfort, R.L.M. | | Deposit date: | 2023-03-23 | | Release date: | 2023-07-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Degron Blocking Strategy Towards Improved CRL4 CRBN Recruiting PROTAC Selectivity.
Chembiochem, 24, 2023
|
|
8OJH
 
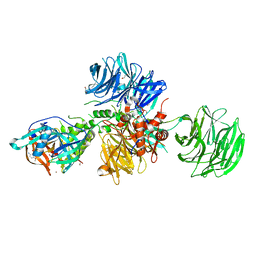 | | Crystal structure of human CRBN-DDB1 in complex with compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 4-azanyl-2-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-7-methoxy-isoindole-1,3-dione, DNA damage-binding protein 1, ... | | Authors: | Cabry, M.P, Le Bihan, Y.-V, van Montfort, R.L.M. | | Deposit date: | 2023-03-24 | | Release date: | 2023-07-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A Degron Blocking Strategy Towards Improved CRL4 CRBN Recruiting PROTAC Selectivity.
Chembiochem, 24, 2023
|
|
6VO1
 
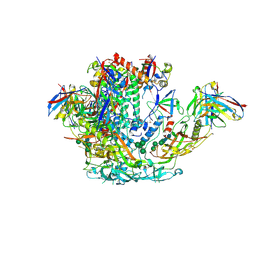 | | BG505 SOSIP.v5.2 in complex with rhesus macaque Fab RM20J | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp120, ... | | Authors: | Cottrell, C.A, Ward, A.B. | | Deposit date: | 2020-01-29 | | Release date: | 2020-07-01 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates.
Plos Pathog., 16, 2020
|
|
6VLR
 
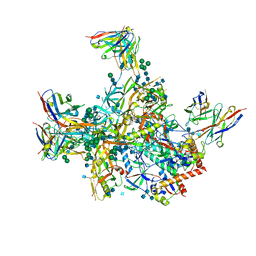 | | BG505 SOSIP.v5.2 in complex with rhesus macaque Fab RM20E1 and PGT122 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BG505 SOSIPv5.2 gp41, ... | | Authors: | Cottrell, C.A, Ward, A.B. | | Deposit date: | 2020-01-24 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.42 Å) | | Cite: | Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates.
Plos Pathog., 16, 2020
|
|
6VN0
 
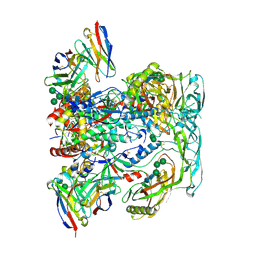 | | BG505 SOSIP.v4.1 in complex with rhesus macaque Fab RM20F | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein gp160, ... | | Authors: | Cottrell, C.A, Shin, M, Ward, A.B. | | Deposit date: | 2020-01-29 | | Release date: | 2020-06-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Mapping the immunogenic landscape of near-native HIV-1 envelope trimers in non-human primates.
Plos Pathog., 16, 2020
|
|
4L62
 
 | |
5ML5
 
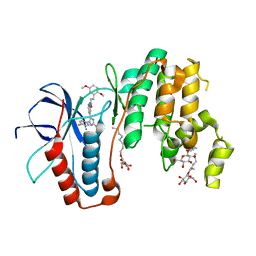 | | Human p38alpha MAPK in complex with imidazolyl pyridine inhibitor 11b | | Descriptor: | 3-(2,5-dimethoxyphenyl)-~{N}-[4-[4-(4-fluorophenyl)-2-methylsulfanyl-1~{H}-imidazol-5-yl]pyridin-2-yl]propanamide, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Buehrmann, M, Rauh, D. | | Deposit date: | 2016-12-06 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Optimized 4,5-Diarylimidazoles as Potent/Selective Inhibitors of Protein Kinase CK1 delta and Their Structural Relation to p38 alpha MAPK.
Molecules, 22, 2017
|
|
5MO4
 
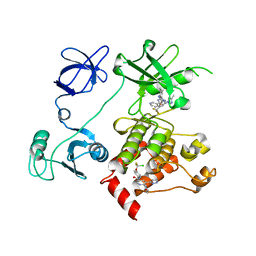 | |
5MQV
 
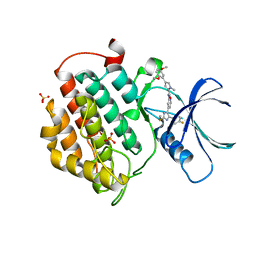 | | Crystal structure of human Casein Kinase I delta in complex with 4-(2,5-Dimethoxyphenyl)-N-(4-(5-(4-fluorphenyl)-2-(methylthio)-1H-imidazol-4-yl)-pyridin-2-yl)-1-methyl-1H-pyrrole-2-carboxamide | | Descriptor: | 4-(2,5-Dimethoxyphenyl)-N-(4-(5-(4-fluorphenyl)-2-(methylthio)-1H-imidazol-4-yl)-pyridin-2-yl)-1-methyl-1H-pyrrole-2-carboxamide, Casein kinase I isoform delta, PHOSPHATE ION | | Authors: | Pichlo, C, Brunstein, E, Baumann, U. | | Deposit date: | 2016-12-20 | | Release date: | 2017-04-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.154 Å) | | Cite: | Optimized 4,5-Diarylimidazoles as Potent/Selective Inhibitors of Protein Kinase CK1 delta and Their Structural Relation to p38 alpha MAPK.
Molecules, 22, 2017
|
|
7U0K
 
 | | IOMA class antibody Fab ACS124 | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, IOMA Class antibody ACS124 Heavy chain, IOMA Class antibody ACS124 Light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
7U04
 
 | | IOMA class antibody ACS101 | | Descriptor: | GLYCEROL, IOMA class antibody ACS101 heavy chain, IOMA class antibody ACS101 light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-02-17 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Identification of IOMA-class neutralizing antibodies targeting the CD4-binding site on the HIV-1 envelope glycoprotein.
Nat Commun, 13, 2022
|
|
1TZE
 
 | |
7U5G
 
 | | ACS122 Fab | | Descriptor: | ACS122 Fab Heavy chain, ACS122 Fab Light chain | | Authors: | Farokhi, E, Stanfield, R.L, Wilson, I.A. | | Deposit date: | 2022-03-02 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Complementary antibody lineages achieve neutralization breadth in an HIV-1 infected elite neutralizer.
Plos Pathog., 18, 2022
|
|
5GGS
 
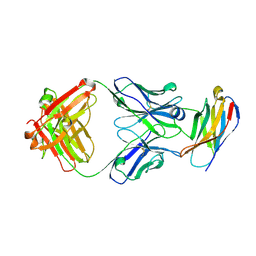 | | PD-1 in complex with pembrolizumab Fab | | Descriptor: | Programmed cell death protein 1, heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
5GGV
 
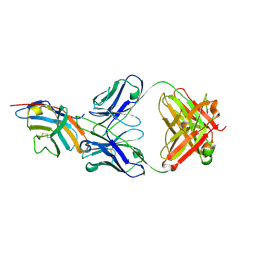 | | CTLA-4 in complex with tremelimumab Fab | | Descriptor: | Cytotoxic T-lymphocyte protein 4, heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
5GGT
 
 | | PD-L1 in complex with BMS-936559 Fab | | Descriptor: | IGK@ protein, IgG H chain, Programmed cell death 1 ligand 1 | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
6BSU
 
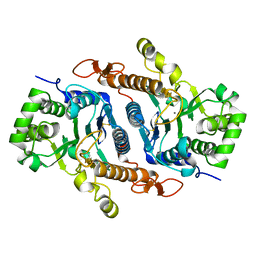 | | Crystal structure of xyloglucan xylosyltransferase I | | Descriptor: | MANGANESE (II) ION, Xyloglucan 6-xylosyltransferase 1 | | Authors: | Culbertson, A.T, Ehrlich, J.J, Choe, J, Honzatko, R.B, Zabotina, O.A. | | Deposit date: | 2017-12-04 | | Release date: | 2018-05-23 | | Last modified: | 2022-03-23 | | Method: | X-RAY DIFFRACTION (1.497 Å) | | Cite: | Structure of xyloglucan xylosyltransferase 1 reveals simple steric rules that define biological patterns of xyloglucan polymers.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BSW
 
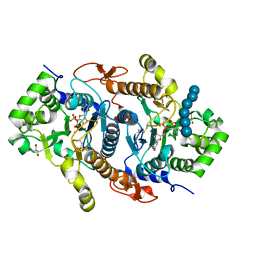 | | Crystal structure of Xyloglucan Xylosyltransferase 1 ternary form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Culbertson, A.T, Ehrlich, J.J, Choe, J, Honzatko, R.B, Zabotina, O.A. | | Deposit date: | 2017-12-04 | | Release date: | 2018-05-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.156 Å) | | Cite: | Structure of xyloglucan xylosyltransferase 1 reveals simple steric rules that define biological patterns of xyloglucan polymers.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BSV
 
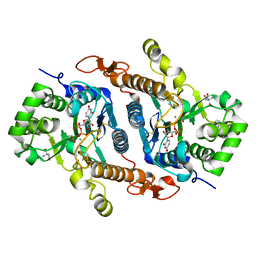 | | Crystal structure of Xyloglucan Xylosyltransferase binary form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, NITRATE ION, ... | | Authors: | Zabotina, O.A, Culbertson, A.T, Ehrlich, J.J, Choe, J, Honzatko, R.B. | | Deposit date: | 2017-12-04 | | Release date: | 2018-05-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.433 Å) | | Cite: | Structure of xyloglucan xylosyltransferase 1 reveals simple steric rules that define biological patterns of xyloglucan polymers.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5GGR
 
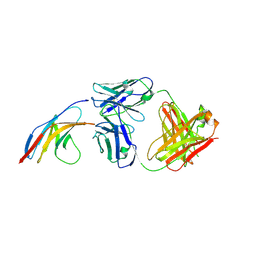 | | PD-1 in complex with nivolumab Fab | | Descriptor: | Programmed cell death protein 1, heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
5GGQ
 
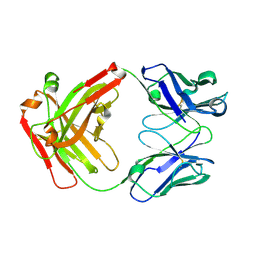 | | Crystal structure of Nivolumab Fab fragment | | Descriptor: | nivolumab heavy chain, nivolumab light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
5VAN
 
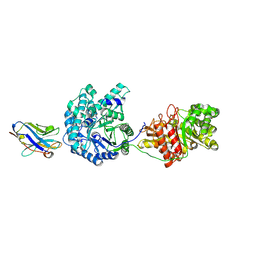 | | Crystal Structure of Beta-Klotho | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lee, S, Schlessinger, J. | | Deposit date: | 2017-03-27 | | Release date: | 2018-01-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Structures of beta-klotho reveal a 'zip code'-like mechanism for endocrine FGF signalling.
Nature, 553, 2018
|
|
5VAK
 
 | | Crystal Structure of Beta-Klotho, Domain 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-klotho, ... | | Authors: | Lee, S, Schlessinger, J. | | Deposit date: | 2017-03-27 | | Release date: | 2018-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of beta-klotho reveal a 'zip code'-like mechanism for endocrine FGF signalling.
Nature, 553, 2018
|
|
5GGU
 
 | | Crystal structure of tremelimumab Fab | | Descriptor: | heavy chain, light chain | | Authors: | Heo, Y.S. | | Deposit date: | 2016-06-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.292 Å) | | Cite: | Structural basis of checkpoint blockade by monoclonal antibodies in cancer immunotherapy
Nat Commun, 7, 2016
|
|
5VAQ
 
 | | Crystal Structure of Beta-Klotho in Complex with FGF21CT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-klotho, Fibroblast growth factor 21, ... | | Authors: | Lee, S, Schlessinger, J. | | Deposit date: | 2017-03-27 | | Release date: | 2018-01-31 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (2.606 Å) | | Cite: | Structures of beta-klotho reveal a 'zip code'-like mechanism for endocrine FGF signalling.
Nature, 553, 2018
|
|
