6LN4
 
 | | Estrogen-related receptor beta(ERR2) in complex with PGC1a-2a | | Descriptor: | 10-mer from Peroxisome proliferator-activated receptor gamma coactivator 1-alpha, Steroid hormone receptor ERR2 | | Authors: | Yao, B.Q, Li, Y. | | Deposit date: | 2019-12-28 | | Release date: | 2020-10-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural Insights into the Specificity of Ligand Binding and Coactivator Assembly by Estrogen-Related Receptor beta.
J.Mol.Biol., 432, 2020
|
|
5NMI
 
 | | Cytochrome bc1 bound to the inhibitor MJM170 | | Descriptor: | (4aS)-2-methyl-3-(4-phenoxyphenyl)-5,6,7,8-tetrahydroquinolin-4(4aH)-one, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, ARG-ASN-TRP-VAL-PRO-THR-ALA-GLN-LEU-TRP-GLY-ALA-VAL-GLY-ALA-VAL-GLY-LEU-VAL-SER-ALA-THR, ... | | Authors: | Capper, N.J, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2017-04-05 | | Release date: | 2017-06-14 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | New paradigms for understanding and step changes in treating active and chronic, persistent apicomplexan infections.
Sci Rep, 6, 2016
|
|
6LTK
 
 | | HSP90 in complex with SNX-2112 | | Descriptor: | 4-[6,6-dimethyl-4-oxidanylidene-3-(trifluoromethyl)-5,7-dihydroindazol-1-yl]-2-[(4-oxidanylcyclohexyl)amino]benzamide, Heat shock protein HSP 90-alpha | | Authors: | Cao, H.L. | | Deposit date: | 2020-01-22 | | Release date: | 2021-01-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Complex Crystal Structure Determination and in vitro Anti-non-small Cell Lung Cancer Activity of Hsp90 N Inhibitor SNX-2112.
Front Cell Dev Biol, 9, 2021
|
|
6L42
 
 | |
8PN9
 
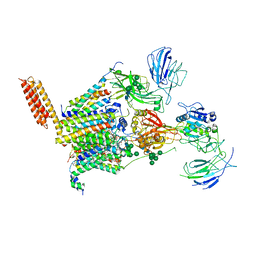 | | Structure of human oligosaccharyltransferase OST-A complex bound to NGI-1 | | Descriptor: | (2E,6E,10E,14E,18E,22E,26E)-3,7,11,15,19,23,27,31-OCTAMETHYLDOTRIACONTA-2,6,10,14,18,22,26,30-OCTAENYL TRIHYDROGEN DIPHOSPHATE, (2~{S},3~{R},4~{R},5~{S},6~{S})-2-(hydroxymethyl)-6-[(1~{S},2~{R},3~{R},4~{R},5'~{S},6~{S},7~{R},8~{S},9~{R},12~{R},13~{R},15~{S},16~{S},18~{R})-5',7,9,13-tetramethyl-3,15-bis(oxidanyl)spiro[5-oxapentacyclo[10.8.0.0^{2,9}.0^{4,8}.0^{13,18}]icosane-6,2'-oxane]-16-yl]oxy-oxane-3,4,5-triol, (4R,7R)-4-hydroxy-N,N,N-trimethyl-4,9-dioxo-7-[(undecanoyloxy)methyl]-3,5,8-trioxa-4lambda~5~-phosphadocosan-1-aminium, ... | | Authors: | Ramirez, A.S, Kowal, J, Locher, K.P. | | Deposit date: | 2023-06-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Positive selection CRISPR screens reveal a druggable pocket in an oligosaccharyltransferase required for inflammatory signaling to NF-kappa B.
Cell, 187, 2024
|
|
5U8F
 
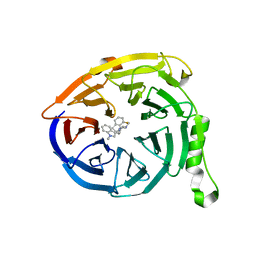 | | Polycomb protein EED in complex with inhibitor: (3R,4S)-1-[(1S)-7-fluoro-2,3-dihydro-1H-inden-1-yl]-N,N-dimethyl-4-(1-methyl-1H-indol-3-yl)pyrrolidin-3-amine | | Descriptor: | (3R,4S)-1-[(1S)-7-fluoro-2,3-dihydro-1H-inden-1-yl]-N,N-dimethyl-4-(1-methyl-1H-indol-3-yl)pyrrolidin-3-amine, Polycomb protein EED | | Authors: | Jakob, C.G, Zhu, H. | | Deposit date: | 2016-12-14 | | Release date: | 2017-03-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.343 Å) | | Cite: | SAR of amino pyrrolidines as potent and novel protein-protein interaction inhibitors of the PRC2 complex through EED binding.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5U69
 
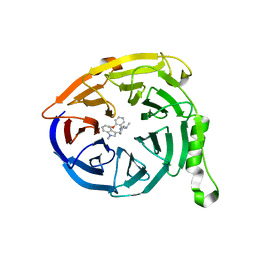 | | Polycomb protein EED in complex with inhibitor: (3R,4S)-1-[(2-methoxyphenyl)methyl]-N,N-dimethyl-4-(1-methylindol-3-yl)pyrrolidin-3-amine | | Descriptor: | (3R,4S)-1-[(2-methoxyphenyl)methyl]-N,N-dimethyl-4-(1-methylindol-3-yl)pyrrolidin-3-amine, Polycomb protein EED | | Authors: | Jakob, C.G, Zhu, H. | | Deposit date: | 2016-12-07 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | SAR of amino pyrrolidines as potent and novel protein-protein interaction inhibitors of the PRC2 complex through EED binding.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5U2M
 
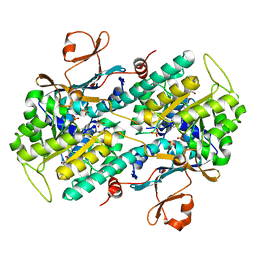 | | Crystal structure of human NAMPT with A-1293201 | | Descriptor: | N-[4-({[(3S)-oxolan-3-yl]methyl}carbamoyl)phenyl]-1,3-dihydro-2H-isoindole-2-carboxamide, Nicotinamide phosphoribosyltransferase, SULFATE ION | | Authors: | Longenecker, K.L, Raich, D, Korepanova, A.V. | | Deposit date: | 2016-11-30 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Discovery and Characterization of Novel Nonsubstrate and Substrate NAMPT Inhibitors.
Mol. Cancer Ther., 16, 2017
|
|
5U8A
 
 | | Polycomb protein EED in complex with inhibitor: (3R,4S)-1-[(2-bromo-6-fluorophenyl)methyl]-N,N-dimethyl-4-(1-methyl-1H-indol-3-yl)pyrrolidin-3-amine | | Descriptor: | (3R,4S)-1-[(2-bromo-6-fluorophenyl)methyl]-N,N-dimethyl-4-(1-methyl-1H-indol-3-yl)pyrrolidin-3-amine, Polycomb protein EED | | Authors: | Jakob, C.G, Zhu, H. | | Deposit date: | 2016-12-14 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | SAR of amino pyrrolidines as potent and novel protein-protein interaction inhibitors of the PRC2 complex through EED binding.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
9J3Z
 
 | | Cryo-EM structure of lysosome cholesterol sencing protein in L state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosomal cholesterol signaling protein,G protein-coupled receptor 155 fusion protein | | Authors: | Qian, H.W, Wang, Z.H. | | Deposit date: | 2024-08-08 | | Release date: | 2025-04-02 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for cholesterol sensing of LYCHOS and its interaction with indoxyl sulfate.
Nat Commun, 16, 2025
|
|
9J3X
 
 | | Cryo-EM structure of lysosome cholesterol sencing protein LYCHOS in Tight-state | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL, ... | | Authors: | Qian, H.W, Wang, Z.H. | | Deposit date: | 2024-08-08 | | Release date: | 2025-04-02 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for cholesterol sensing of LYCHOS and its interaction with indoxyl sulfate.
Nat Commun, 16, 2025
|
|
9J40
 
 | | Cryo-EM structure of lysosome cholesterol sencing protein complex | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-SULFOOXY-1H-INDOLE, ... | | Authors: | Qian, H.W, Wang, Z.H. | | Deposit date: | 2024-08-08 | | Release date: | 2025-04-02 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for cholesterol sensing of LYCHOS and its interaction with indoxyl sulfate.
Nat Commun, 16, 2025
|
|
8E6Y
 
 | |
2MVT
 
 | | Solution structure of scoloptoxin SSD609 from Scolopendra mutilans | | Descriptor: | Scoloptoxin SSD609 | | Authors: | Wu, F, Sun, P, Wang, C, He, Y, Zhang, L, Tian, C. | | Deposit date: | 2014-10-14 | | Release date: | 2015-09-23 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | A distinct three-helix centipede toxin SSD609 inhibits Iks channels by interacting with the KCNE1 auxiliary subunit.
Sci Rep, 5, 2015
|
|
6L8O
 
 | | Crystal structure of the K. lactis Rad5 (Hg-derivative) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA repair protein RAD5, MERCURY (II) ION | | Authors: | Shen, M, Xiang, S. | | Deposit date: | 2019-11-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for the multi-activity factor Rad5 in replication stress tolerance.
Nat Commun, 12, 2021
|
|
6L8N
 
 | | Crystal structure of the K. lactis Rad5 | | Descriptor: | DNA repair protein RAD5, ZINC ION | | Authors: | Shen, M, Xiang, S. | | Deposit date: | 2019-11-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis for the multi-activity factor Rad5 in replication stress tolerance.
Nat Commun, 12, 2021
|
|
6UAI
 
 | | Imidazole-triggered RAS-specific subtilisin SUBT_BACAM complexed with YSAM peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Toth, E.A, Bryan, P.N, Orban, J. | | Deposit date: | 2019-09-10 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Engineering subtilisin proteases that specifically degrade active RAS.
Commun Biol, 4, 2021
|
|
6UAO
 
 | | Imidazole-triggered RAS-specific subtilisin SUBT_BACAM complexed with the peptide EEYSAM | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peptide EEYSAM, ... | | Authors: | Toth, E.A, Bryan, P.N, Orban, J. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Engineering subtilisin proteases that specifically degrade active RAS.
Commun Biol, 4, 2021
|
|
6UBE
 
 | | Azide-triggered subtilisin SUBT_BACAM complexed with the peptide LFRAL | | Descriptor: | AZIDE ION, GLYCEROL, Peptide LFRAL, ... | | Authors: | Toth, E.A, Bryan, P.N, Orban, J, Gallagher, D.T, Custer, G. | | Deposit date: | 2019-09-11 | | Release date: | 2020-09-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Engineering subtilisin proteases that specifically degrade active RAS.
Commun Biol, 4, 2021
|
|
3L3Z
 
 | | Crystal structure of DHT-bound androgen receptor in complex with the third motif of steroid receptor coactivator 3 | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Nuclear receptor coactivator 3 | | Authors: | Zhou, X.E, Suino-Powell, K.M, Li, J, He, A, MacKeigan, J.P, Melcher, K, Yong, E.-L, Xu, H.E. | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of SRC3/AIB1 as a Preferred Coactivator for Hormone-activated Androgen Receptor.
J.Biol.Chem., 285, 2010
|
|
8GYB
 
 | |
8GYA
 
 | |
8GY4
 
 | | Crystal structure of Alongshan virus methyltransferase | | Descriptor: | Methyltransferase | | Authors: | Chen, H, Lin, S, Lu, G.W. | | Deposit date: | 2022-09-21 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and functional basis of low-affinity SAM/SAH-binding in the conserved MTase of the multi-segmented Alongshan virus distantly related to canonical unsegmented flaviviruses.
Plos Pathog., 19, 2023
|
|
8GY9
 
 | |
3L3X
 
 | | Crystal structure of DHT-bound androgen receptor in complex with the first motif of steroid receptor coactivator 3 | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Nuclear receptor coactivator 3 | | Authors: | Zhou, X.E, Suino-Powell, K.M, Li, J, He, A, MacKeigan, J.P, Melcher, K, Yong, E.-L, Xu, H.E. | | Deposit date: | 2009-12-18 | | Release date: | 2010-01-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of SRC3/AIB1 as a Preferred Coactivator for Hormone-activated Androgen Receptor.
J.Biol.Chem., 285, 2010
|
|
