2ZVO
 
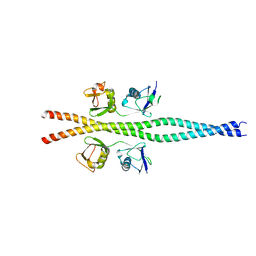 | | NEMO CoZi domain in complex with diubiquitin in C2 space group | | Descriptor: | NF-kappa-B essential modulator, UBC protein | | Authors: | Rahighi, S, Ikeda, F, Kawasaki, M, Akutsu, M, Suzuki, N, Kato, R, Kensche, T, Uejima, T, Bloor, S, Komander, D, Randow, F, Wakatsuki, S, Dikic, I. | | Deposit date: | 2008-11-12 | | Release date: | 2009-03-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation
Cell(Cambridge,Mass.), 136, 2009
|
|
2N3E
 
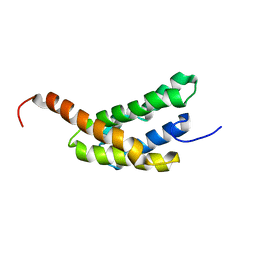 | | Amino-terminal domain of Latrodectus hesperus MaSp1 with neutralized acidic cluster | | Descriptor: | Major ampullate spidroin 1 | | Authors: | Schaal, D, Bauer, J, Schweimer, K, Scheibel, T, Roesch, P, Schwarzinger, S. | | Deposit date: | 2015-05-29 | | Release date: | 2016-06-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | High resolution structure of an engineered amino-terminal ampullate spider silk with neutralized charge cluster
To be Published
|
|
2ZVN
 
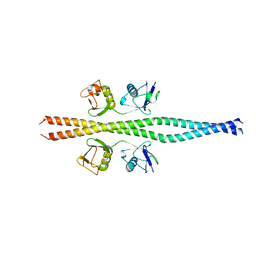 | | NEMO CoZi domain incomplex with diubiquitin in P212121 space group | | Descriptor: | NF-kappa-B essential modulator, UBC protein | | Authors: | Rahighi, S, Ikeda, F, Kawasaki, M, Akutsu, M, Suzuki, N, Kato, R, Kensche, T, Uejima, T, Bloor, S, Komander, D, Randow, F, Wakatsuki, S, Dikic, I. | | Deposit date: | 2008-11-12 | | Release date: | 2009-03-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation
Cell(Cambridge,Mass.), 136, 2009
|
|
3F89
 
 | | NEMO CoZi domain | | Descriptor: | NF-kappa-B essential modulator | | Authors: | Rahighi, S, Ikeda, F, Kawasaki, M, Akutsu, M, Suzuki, N, Kato, R, Kensche, T, Uejima, T, Bloor, S, Komander, D, Randow, F, Wakatsuki, S, Dikic, I. | | Deposit date: | 2008-11-11 | | Release date: | 2009-03-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation
Cell(Cambridge,Mass.), 136, 2009
|
|
3PR2
 
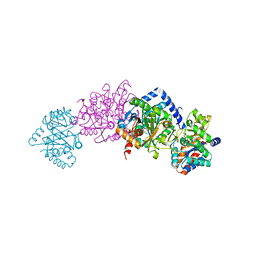 | | Tryptophan synthase indoline quinonoid structure with F9 inhibitor in alpha site | | Descriptor: | (Z)-N-[(1E)-1-carboxy-2-(2,3-dihydro-1H-indol-1-yl)ethylidene]{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4(1H)-ylidene}methanaminium, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, CESIUM ION, ... | | Authors: | Lai, J, Niks, D, Wang, Y, Domratcheva, T, Barends, T.R.M, Schwarz, F, Olsen, R.A, Elliott, D.W, Fatmi, M.Q, Chang, C.A, Schlichting, I, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2010-11-29 | | Release date: | 2011-02-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | X-ray and NMR Crystallography in an Enzyme Active Site: The Indoline Quinonoid Intermediate in Tryptophan Synthase.
J.Am.Chem.Soc., 133, 2011
|
|
4EKW
 
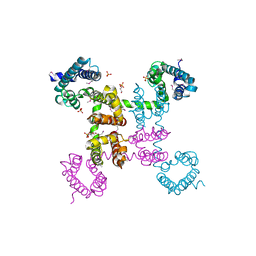 | | Crystal structure of the NavAb voltage-gated sodium channel (wild-type, 3.2 A) | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Ion transport protein, PHOSPHATE ION | | Authors: | Payandeh, J, Gamal El-Din, T.M, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2012-04-10 | | Release date: | 2012-05-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Crystal structure of a voltage-gated sodium channel in two potentially inactivated states.
Nature, 486, 2012
|
|
2VRY
 
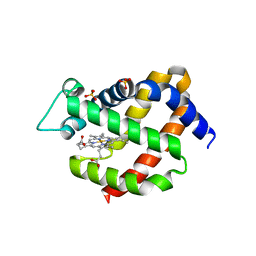 | | Mouse Neuroglobin with heme iron in the reduced ferrous state | | Descriptor: | NEUROGLOBIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Arcovito, A, Moschetti, T, D'Angelo, P, Mancini, G, Vallone, B, Brunori, M, Della Longa, S. | | Deposit date: | 2008-04-16 | | Release date: | 2008-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | An X-Ray Diffraction and X-Ray Absorption Spectroscopy Joint Study of Neuroglobin.
Arch.Biochem.Biophys., 475, 2008
|
|
3RW0
 
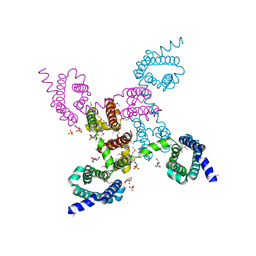 | | Crystal structure of the NavAb voltage-gated sodium channel (Met221Cys, 2.95 A) | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Ion transport protein | | Authors: | Payandeh, J, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2011-05-06 | | Release date: | 2011-07-13 | | Last modified: | 2011-07-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | The crystal structure of a voltage-gated sodium channel.
Nature, 475, 2011
|
|
3RVZ
 
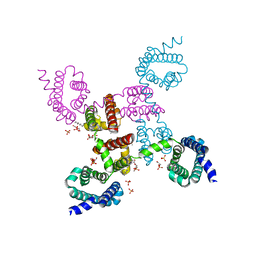 | | Crystal structure of the NavAb voltage-gated sodium channel (Ile217Cys, 2.8 A) | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Ion transport protein | | Authors: | Payandeh, J, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2011-05-06 | | Release date: | 2011-07-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of a voltage-gated sodium channel.
Nature, 475, 2011
|
|
2WON
 
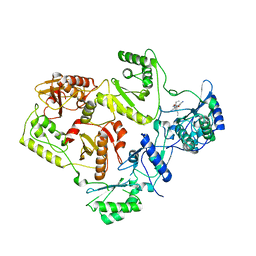 | | Crystal Structure of UK-453061 bound to HIV-1 Reverse Transcriptase (wild-type). | | Descriptor: | 5-{[3,5-diethyl-1-(2-hydroxyethyl)-1H-pyrazol-4-yl]oxy}benzene-1,3-dicarbonitrile, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Phillips, C, Irving, S.L, Knoechel, T, Ringrose, H. | | Deposit date: | 2009-07-27 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Lersivirine: A Non-Nucleoside Reverse Transcriptase Inhibitor with Activity Against Drug- Resistant Human Immunodeficiency Virus-1.
Antimicrob.Agents Chemother., 54, 2010
|
|
2QZR
 
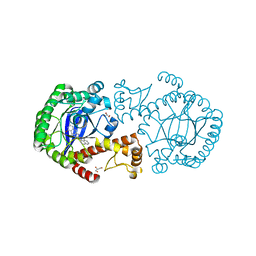 | | tRNA-Guanine Transglycosylase(TGT) in Complex with 6-amino-2-[(1-naphthylmethyl)amino]-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one | | Descriptor: | 6-amino-2-[(1-naphthylmethyl)amino]-3,7-dihydro-8H-imidazo[4,5-g]quinazolin-8-one, GLYCEROL, Queuine tRNA-ribosyltransferase, ... | | Authors: | Hoertner, S.R, Ritschel, T, Stengl, B, Kramer, C, Schweizer, W.B, Wagner, B, Kansy, M, Klebe, G, Diederich, F. | | Deposit date: | 2007-08-17 | | Release date: | 2007-09-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Potent Inhibitors of tRNA-Guanine Transglycosylase, an Enzyme Linked to the Pathogenicity of the Shigella Bacterium: Charge-Assisted Hydrogen Bonding.
Angew.Chem.Int.Ed.Engl., 46, 2007
|
|
3RVY
 
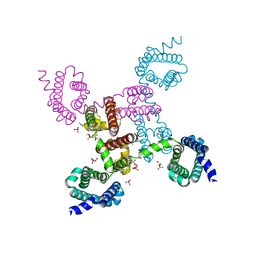 | | Crystal structure of the NavAb voltage-gated sodium channel (Ile217Cys, 2.7 A) | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Ion transport protein | | Authors: | Payandeh, J, Scheuer, T, Zheng, N, Catterall, W.A. | | Deposit date: | 2011-05-06 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The crystal structure of a voltage-gated sodium channel.
Nature, 475, 2011
|
|
2LAG
 
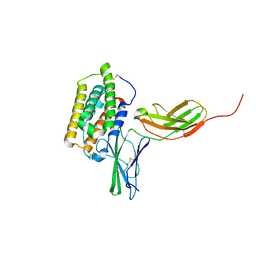 | | Structure of the 44 kDa complex of interferon-alpha2 with the extracellular part of IFNAR2 obtained by 2D-double difference NOESY | | Descriptor: | Interferon alpha-2, Interferon alpha/beta receptor 2 | | Authors: | Nudelman, I, Akabayov, S.R, Scherf, T, Anglister, J. | | Deposit date: | 2011-03-13 | | Release date: | 2011-08-17 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Observation of Intermolecular Interactions in Large Protein Complexes by 2D-Double Difference Nuclear Overhauser Enhancement Spectroscopy: Application to the 44 kDa Interferon-Receptor Complex.
J.Am.Chem.Soc., 133, 2011
|
|
1N9N
 
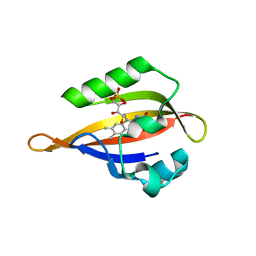 | | Crystal structure of the Phot-LOV1 domain from Chlamydomonas reinhardtii in illuminated state. Data set of a single crystal. | | Descriptor: | FLAVIN MONONUCLEOTIDE, putative blue light receptor | | Authors: | Fedorov, R, Schlichting, I, Hartmann, E, Domratcheva, T, Fuhrmann, M, Hegemann, P. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures and molecular mechanism of a light-induced signaling switch: The Phot-LOV1 domain from Chlamydomonas reinhardtii.
Biophys.J., 84, 2003
|
|
3C09
 
 | | Crystal structure the Fab fragment of matuzumab (Fab72000) in complex with domain III of the extracellular region of EGFR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Epidermal growth factor receptor, Matuzumab Fab Heavy chain, ... | | Authors: | Ferguson, K.M, Schmiedel, J, Knoechel, T. | | Deposit date: | 2008-01-18 | | Release date: | 2008-04-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Matuzumab binding to EGFR prevents the conformational rearrangement required for dimerization.
Cancer Cell, 13, 2008
|
|
3C08
 
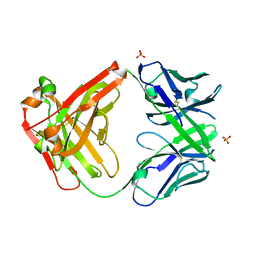 | |
1N9O
 
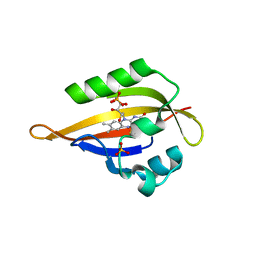 | | Crystal structure of the Phot-LOV1 domain from Chlamydomonas reinhardtii in illuminated state. Composite data set. | | Descriptor: | FLAVIN MONONUCLEOTIDE, SULFATE ION, putative blue light receptor | | Authors: | Fedorov, R, Schlichting, I, Hartmann, E, Domratcheva, T, Fuhrmann, M, Hegemann, P. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures and molecular mechanism of a light-induced signaling switch: The Phot-LOV1 domain from Chlamydomonas reinhardtii.
Biophys.J., 84, 2003
|
|
2ZJP
 
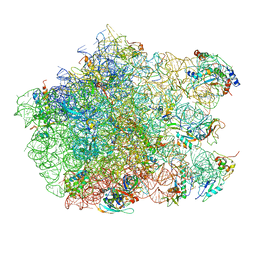 | | Thiopeptide antibiotic Nosiheptide bound to the large ribosomal subunit of Deinococcus radiodurans | | Descriptor: | 4-(hydroxymethyl)-3-methyl-1H-indole-2-carboxylic acid, 50S RIBOSOMAL PROTEIN L11, 50S RIBOSOMAL PROTEIN L13, ... | | Authors: | Harms, J.M, Wilson, D.N, Schluenzen, F, Connell, S.R, Stachelhaus, T, Zaborowska, Z, Spahn, C.M.T, Fucini, P. | | Deposit date: | 2008-03-07 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Translational Regulation Via L11: Molecular Switches on the Ribosome Turned on and Off by Thiostrepton and Micrococcin.
Mol.Cell, 30, 2008
|
|
2WOM
 
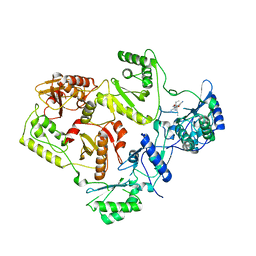 | | Crystal Structure of UK-453061 bound to HIV-1 Reverse Transcriptase (K103N). | | Descriptor: | 5-{[3,5-diethyl-1-(2-hydroxyethyl)-1H-pyrazol-4-yl]oxy}benzene-1,3-dicarbonitrile, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Phillips, C, Irving, S.L, Knoechel, T, Ringrose, H. | | Deposit date: | 2009-07-27 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Lersivirine, a nonnucleoside reverse transcriptase inhibitor with activity against drug-resistant human immunodeficiency virus type 1.
Antimicrob. Agents Chemother., 54, 2010
|
|
3IF9
 
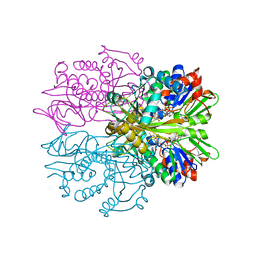 | | Crystal structure of Glycine Oxidase G51S/A54R/H244A mutant in complex with inhibitor glycolate | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCOLIC ACID, Glycine oxidase | | Authors: | Pedotti, M, Rosini, E, Molla, G, Moschetti, T, Vallone, B, Savino, C, Pollegioni, L. | | Deposit date: | 2009-07-24 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Glyphosate resistance by engineering the flavoenzyme glycine oxidase.
J.Biol.Chem., 284, 2009
|
|
1N9L
 
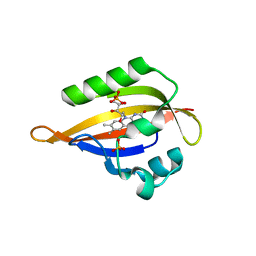 | | Crystal structure of the Phot-LOV1 domain from Chlamydomonas reinhardtii in the dark state. | | Descriptor: | FLAVIN MONONUCLEOTIDE, SULFATE ION, putative blue light receptor | | Authors: | Fedorov, R, Schlichting, I, Hartmann, E, Domratcheva, T, Fuhrmann, M, Hegemann, P. | | Deposit date: | 2002-11-25 | | Release date: | 2003-06-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures and molecular mechanism of a light-induced signaling switch: The Phot-LOV1 domain from Chlamydomonas reinhardtii.
Biophys.J., 84, 2003
|
|
2ZJQ
 
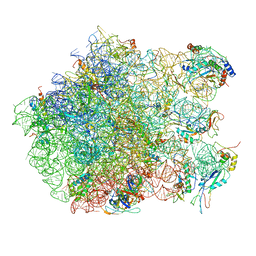 | | Interaction of L7 with L11 induced by Microccocin binding to the Deinococcus radiodurans 50S subunit | | Descriptor: | 50S ribosomal protein L11, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Harms, J.M, Wilson, D.N, Schluenzen, F, Connell, S.R, Stachelhaus, T, Zaborowska, Z, Spahn, C.M.T, Fucini, P. | | Deposit date: | 2008-03-08 | | Release date: | 2008-06-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin.
Mol.Cell, 30, 2008
|
|
2ZJR
 
 | | Refined native structure of the large ribosomal subunit (50S) from Deinococcus radiodurans | | Descriptor: | 50S ribosomal protein L11, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Harms, J.M, Wilson, D.N, Schluenzen, F, Connell, S.R, Stachelhaus, T, Zaborowska, Z, Spahn, C.M.T, Fucini, P. | | Deposit date: | 2008-03-08 | | Release date: | 2008-06-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin.
Mol.Cell, 30, 2008
|
|
1R0E
 
 | | Glycogen synthase kinase-3 beta in complex with 3-indolyl-4-arylmaleimide inhibitor | | Descriptor: | 3-[3-(2,3-DIHYDROXY-PROPYLAMINO)-PHENYL]-4-(5-FLUORO-1-METHYL-1H-INDOL-3-YL)-PYRROLE-2,5-DIONE, CITRATE ANION, Glycogen synthase kinase-3 beta | | Authors: | Allard, J, Nikolcheva, T, Gong, L, Wang, J, Dunten, P, Avnur, Z, Waters, R, Sun, Q, Skinner, B. | | Deposit date: | 2003-09-20 | | Release date: | 2004-10-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | From genetics to therapeutics: the Wnt pathway and osteoporosis
To be Published
|
|
2GIF
 
 | | Asymmetric structure of trimeric AcrB from Escherichia coli | | Descriptor: | Acriflavine resistance protein B, CITRATE ANION | | Authors: | Seeger, M.A, Schiefner, A, Eicher, T, Verrey, F, Diederichs, K, Pos, K.M. | | Deposit date: | 2006-03-28 | | Release date: | 2006-09-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural Asymmetry of AcrB Trimer Suggests a Peristaltic Pump Mechanism.
Science, 313, 2006
|
|
