1M3G
 
 | |
6DJC
 
 | | Crystal structure of human Bromodomain-containing protein 4 (BRD4) bromodomain with MS645 | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, N,N'-(decane-1,10-diyl)bis{2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetamide} | | Authors: | Ren, C, Zhou, M.M. | | Deposit date: | 2018-05-25 | | Release date: | 2018-07-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Spatially constrained tandem bromodomain inhibition bolsters sustained repression of BRD4 transcriptional activity for TNBC cell growth.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
7BVH
 
 | | Crystal structure of arabinosyltransferase EmbC2-AcpM2 complex from Mycobacterium smegmatis complexed with di-arabinose | | Descriptor: | CALCIUM ION, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, Meromycolate extension acyl carrier protein, ... | | Authors: | Zhao, Y, Zhang, L, Wu, L.J, Wang, Q, Li, J, Besra, G.S, Rao, Z.H. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVF
 
 | | Cryo-EM structure of Mycobacterium tuberculosis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | | Descriptor: | CALCIUM ION, CARDIOLIPIN, Ethambutol, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVC
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with ethambutol | | Descriptor: | 4'-PHOSPHOPANTETHEINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVE
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbC2-AcpM2 in complex with ethambutol | | Descriptor: | CALCIUM ION, Ethambutol, Integral membrane indolylacetylinositol arabinosyltransferase EmbC, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2020-07-01 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
7BVG
 
 | | Cryo-EM structure of Mycobacterium smegmatis arabinosyltransferase EmbA-EmbB-AcpM2 in complex with di-arabinose. | | Descriptor: | 4'-PHOSPHOPANTETHEINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhang, L, Zhao, Y, Gao, Y, Wang, Q, Li, J, Besra, G.S, Rao, Z. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-29 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of cell wall arabinosyltransferases with the anti-tuberculosis drug ethambutol.
Science, 368, 2020
|
|
2QMG
 
 | | Structure of BACE Bound to SCH745966 | | Descriptor: | Beta-secretase 1, D(-)-TARTARIC ACID, N-{(1S,2R)-1-(3,5-DIFLUOROBENZYL)-2-HYDROXY-2-[(2R,4R)-4-PHENOXYPYRROLIDIN-2-YL]ETHYL}-3-{[(2R)-2-(METHOXYMETHYL)PYRROLIDIN-1-YL]CARBONYL}-5-METHYLBENZAMIDE | | Authors: | Strickland, C.O, Iserloh, U. | | Deposit date: | 2007-07-16 | | Release date: | 2008-03-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Discovery of an orally efficaceous 4-phenoxypyrrolidine-based BACE-1 inhibitor.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
6K9O
 
 | | Crystal Structure Analysis of Protein | | Descriptor: | Endo-1,4-beta-xylanase 2, GLYCEROL, IODIDE ION | | Authors: | Li, C, Wan, Q. | | Deposit date: | 2019-06-17 | | Release date: | 2020-06-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Studying the Role of a Single Mutation of a Family 11 Glycoside Hydrolase Using High-Resolution X-ray Crystallography.
Protein J., 39, 2020
|
|
2GA2
 
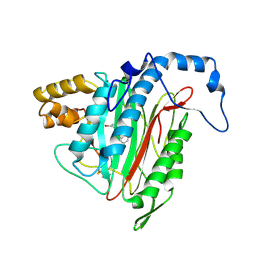 | | h-MetAP2 complexed with A193400 | | Descriptor: | 5-BROMO-2-{[(4-CHLOROPHENYL)SULFONYL]AMINO}BENZOIC ACID, MANGANESE (II) ION, Methionine aminopeptidase 2 | | Authors: | Park, C. | | Deposit date: | 2006-03-07 | | Release date: | 2007-03-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Development of sulfonamide compounds as potent methionine aminopeptidase type II inhibitors with antiproliferative properties.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
7BWJ
 
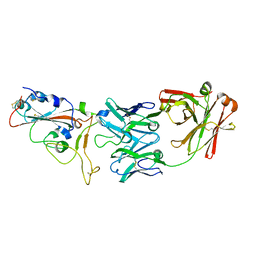 | | crystal structure of SARS-CoV-2 antibody with RBD | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, antibody heavy chain, ... | | Authors: | Wang, X, Ge, J. | | Deposit date: | 2020-04-14 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Human neutralizing antibodies elicited by SARS-CoV-2 infection.
Nature, 584, 2020
|
|
3HGG
 
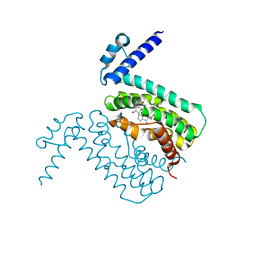 | | Crystal Structure of CmeR Bound to Cholic Acid | | Descriptor: | CHOLIC ACID, CmeR | | Authors: | Routh, M.D, Yang, F. | | Deposit date: | 2009-05-13 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural basis for anionic ligand recognition by multidrug
binding proteins: Crystal structures of CmeR-bile acid complexes
To be Published
|
|
3HGY
 
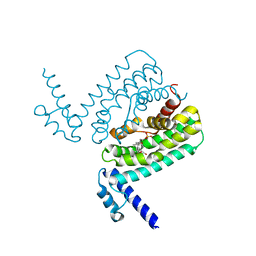 | | Crystal Structure of CmeR Bound to Taurocholic Acid | | Descriptor: | CmeR, TAUROCHOLIC ACID | | Authors: | Routh, M.D, Yang, F. | | Deposit date: | 2009-05-14 | | Release date: | 2010-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.416 Å) | | Cite: | Structural basis for anionic ligand recognition by multidrug
binding proteins: crystal structures of CmeR-bile acid complexes
To be Published
|
|
5DG8
 
 | | CRYSTAL STRUCTURE OF HUMAN DNA POLYMERASE ETA INSERTING dAMPNPP ACROSS A DNA TEMPLATE CONTAINING 1,N6-ETHENODEOXYADENOSINE LESION | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*(EDA)P*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2015-08-27 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural and Kinetic Analysis of Miscoding Opposite the DNA Adduct 1,N6-Ethenodeoxyadenosine by Human Translesion DNA Polymerase eta.
J.Biol.Chem., 291, 2016
|
|
5DG7
 
 | | CRYSTAL STRUCTURE OF HUMAN DNA POLYMERASE ETA INSERTING dTTP ACROSS A DNA TEMPLATE CONTAINING 1,N6-ETHENODEOXYADENOSINE LESION | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*(EDA)P*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2015-08-27 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Structural and Kinetic Analysis of Miscoding Opposite the DNA Adduct 1,N6-Ethenodeoxyadenosine by Human Translesion DNA Polymerase eta.
J.Biol.Chem., 291, 2016
|
|
5DGB
 
 | | CRYSTAL STRUCTURE OF HUMAN DNA POLYMERASE ETA EXTENDING AN 1,N6-ETHENODEOXYADENOSINE : dA PAIR BY INSERTING dTMPNPP OPPOSITE TEMPLATE dA | | Descriptor: | 5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]thymidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*A)-3'), DNA (5'-D(*CP*AP*TP*AP*(EDA)P*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2015-08-27 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural and Kinetic Analysis of Miscoding Opposite the DNA Adduct 1,N6-Ethenodeoxyadenosine by Human Translesion DNA Polymerase eta.
J.Biol.Chem., 291, 2016
|
|
4IGQ
 
 | | Histone H3 Lysine 4 Demethylating Rice JMJ703 in complex with methylated H3K4 substrate | | Descriptor: | FE (III) ION, N-OXALYLGLYCINE, Os05g0196500 protein, ... | | Authors: | Chen, Q.F, Chen, X.S, Wang, Q, Zhang, F.B, Lou, Z.Y, Zhang, Q.F, Zhou, D.X. | | Deposit date: | 2012-12-17 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of a histone H3 lysine 4 demethylase required for stem elongation in rice.
PLoS Genet., 9, 2013
|
|
3KLX
 
 | |
4IGP
 
 | | Histone H3 Lysine 4 Demethylating Rice JMJ703 apo enzyme | | Descriptor: | FE (III) ION, Os05g0196500 protein | | Authors: | Chen, Q.F, Chen, X.S, Wang, Q, Zhang, F.B, Lou, Z.Y, Zhang, Q.F, Zhou, D.X. | | Deposit date: | 2012-12-17 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Structural basis of a histone H3 lysine 4 demethylase required for stem elongation in rice.
PLoS Genet., 9, 2013
|
|
4IGO
 
 | | Histone H3 Lysine 4 Demethylating rice Rice JMJ703 in complex with alpha-KG | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, Os05g0196500 protein | | Authors: | Chen, Q.F, Chen, X.S, Wang, Q, Zhang, F.B, Lou, Z.Y, Zhang, Q.F, Zhou, D.X. | | Deposit date: | 2012-12-17 | | Release date: | 2013-04-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of a histone H3 lysine 4 demethylase required for stem elongation in rice.
PLoS Genet., 9, 2013
|
|
3KL1
 
 | |
6DUV
 
 | | Crystal structure of the second bromodomain of human BRD4 in complex with MS417 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain-containing protein 4, methyl [(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl]acetate | | Authors: | Babault, N, Zhou, M.M. | | Deposit date: | 2018-06-22 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Role of Bound Water Molecules in Differential Recognition of Di-Acetylated Histone Peptides by the BRD4 Bromodomains
To be published
|
|
4O3O
 
 | | Crystal structure of human polymerase eta inserting datp opposite an 8-oxog containing dna template | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]adenosine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*T)-3'), DNA (5'-D(*CP*AP*TP*(8OG)P*AP*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | Kinetics, Structure, and Mechanism of 8-Oxo-7,8-dihydro-2'-deoxyguanosine Bypass by Human DNA Polymerase eta
J.Biol.Chem., 289, 2014
|
|
4O62
 
 | | CW-type zinc finger of ZCWPW2 in complex with the amino terminus of histone H3 | | Descriptor: | Histone H3.3, UNKNOWN ATOM OR ION, ZINC ION, ... | | Authors: | Liu, Y, Tempel, W, Dong, A, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-20 | | Release date: | 2014-03-26 | | Last modified: | 2016-06-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Family-wide Characterization of Histone Binding Abilities of Human CW Domain-containing Proteins.
J.Biol.Chem., 291, 2016
|
|
4O3S
 
 | | Crystal structure of human polymerase eta extending an 8-oxog dna lesion: post insertion of 8-oxog-dc pair | | Descriptor: | 2'-deoxy-5'-O-[(R)-hydroxy{[(R)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]cytidine, DNA (5'-D(*AP*GP*CP*GP*TP*CP*AP*C)-3'), DNA (5'-D(*CP*AP*TP*GP*(8OG)P*TP*GP*AP*CP*GP*CP*T)-3'), ... | | Authors: | Patra, A, Egli, M. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.717 Å) | | Cite: | Kinetics, Structure, and Mechanism of 8-Oxo-7,8-dihydro-2'-deoxyguanosine Bypass by Human DNA Polymerase eta
J.Biol.Chem., 289, 2014
|
|
