8FY9
 
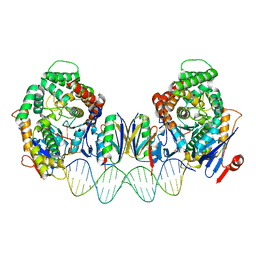 | | Cryo-EM structure of Cas1:Cas2-DEDDh:PAM-deficient prespacer complex | | Descriptor: | Cas1, Cas2-DEDDh, DNA (28-MER) | | Authors: | Skopintsev, P, Tuck, O.T, Soczek, K.M, Doudna, J. | | Deposit date: | 2023-01-25 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Genome expansion by a CRISPR trimmer-integrase.
Nature, 618, 2023
|
|
8FYD
 
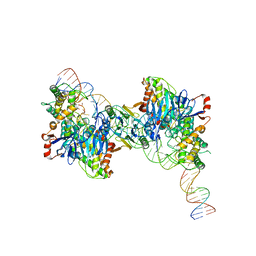 | | Cryo-EM structure of Cas1:Cas2-DEDDh:half-site integration complex bent CRISPR repeat conformation | | Descriptor: | Cas1, Cas2-DEDDh, DNA (13-MER), ... | | Authors: | Skopintsev, P, Tuck, O.T, Soczek, K.M, Doudna, J. | | Deposit date: | 2023-01-25 | | Release date: | 2023-05-03 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Genome expansion by a CRISPR trimmer-integrase.
Nature, 618, 2023
|
|
7YP0
 
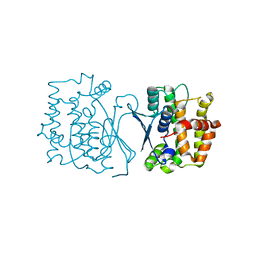 | | Crystal structure of CtGST | | Descriptor: | Glutathione S-transferase | | Authors: | Yang, J, Fan, J.P, Lei, X.G. | | Deposit date: | 2022-08-02 | | Release date: | 2024-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
7YOC
 
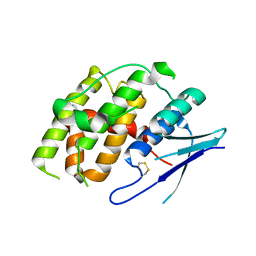 | | Crystal structure of Fhb7 | | Descriptor: | Fhb7 | | Authors: | Yang, J, Liang, K, Xiao, J.Y, Lei, X.G. | | Deposit date: | 2022-08-01 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Enzymatic Degradation of Deoxynivalenol with the Engineered Detoxification Enzyme Fhb7.
Jacs Au, 4, 2024
|
|
8G5T
 
 | | Crystal structure of apo TnmK2 | | Descriptor: | TnmK2 | | Authors: | Liu, Y.-C, Gui, C, Shen, B. | | Deposit date: | 2023-02-14 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.846 Å) | | Cite: | Cofactorless oxygenases guide anthraquinone-fused enediyne biosynthesis.
Nat.Chem.Biol., 20, 2024
|
|
8G5U
 
 | | Crystal structure of TnmK2 complexed with TNM B | | Descriptor: | TnmK2, methyl (2E)-3-[(1aS,11S,11aS,14Z,18R)-3,18-dihydroxy-4,9-dioxo-4,9,10,11-tetrahydro-11aH-11,1a-hept[3]ene[1,5]diynonaphtho[2,3-h]oxireno[c]quinolin-11a-yl]but-2-enoate | | Authors: | Liu, Y.-C, Gui, C, Shen, B. | | Deposit date: | 2023-02-14 | | Release date: | 2023-10-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.804 Å) | | Cite: | Cofactorless oxygenases guide anthraquinone-fused enediyne biosynthesis.
Nat.Chem.Biol., 20, 2024
|
|
8H77
 
 | | Hsp90-AhR-p23-XAP2 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AH receptor-interacting protein, Aryl hydrocarbon receptor, ... | | Authors: | Wen, Z.L, Zhai, Y.J, Zhu, Y, Sun, F. | | Deposit date: | 2022-10-19 | | Release date: | 2023-01-04 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structure of the cytosolic AhR complex.
Structure, 31, 2023
|
|
5H48
 
 | | Crystal structure of Cbln1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-1 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
5H4C
 
 | | Crystal structure of Cbln4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein Cbln4 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
8HO1
 
 | | Crystal structure of cytochrome P450 NasF5053 mutant F387G | | Descriptor: | CALCIUM ION, Cytochrome P450-F5053, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ma, B.D, Tian, W, Qu, X, Kong, X.D. | | Deposit date: | 2022-12-09 | | Release date: | 2023-04-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering the Substrate Specificity of a P450 Dimerase Enables the Collective Biosynthesis of Heterodimeric Tryptophan-Containing Diketopiperazines.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HNZ
 
 | | Crystal structure of cytochrome P450 NasF5053 mutant E73S complexed with 6FCWP | | Descriptor: | (3~{S},8~{a}~{S})-3-[(5-fluoranyl-1~{H}-indol-3-yl)methyl]-2,3,6,7,8,8~{a}-hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, CALCIUM ION, Cytochrome P450-F5053, ... | | Authors: | Ma, B.D, Tian, W, Qu, X, Kong, X.D. | | Deposit date: | 2022-12-09 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Engineering the Substrate Specificity of a P450 Dimerase Enables the Collective Biosynthesis of Heterodimeric Tryptophan-Containing Diketopiperazines.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HO0
 
 | | Crystal structure of cytochrome P450 NasF5053 mutant E73S complexed with 8FCWP | | Descriptor: | (3~{S},8~{a}~{S})-3-[(7-fluoranyl-1~{H}-indol-3-yl)methyl]-2,3,6,7,8,8~{a}-hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, CALCIUM ION, Cytochrome P450-F5053, ... | | Authors: | Ma, B.D, Tian, W, Qu, X, Kong, X.D. | | Deposit date: | 2022-12-09 | | Release date: | 2023-04-19 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Engineering the Substrate Specificity of a P450 Dimerase Enables the Collective Biosynthesis of Heterodimeric Tryptophan-Containing Diketopiperazines.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HNY
 
 | | Crystal structure of cytochrome P450 NasF5053 mutant E73S complexed with 5FCWP | | Descriptor: | (3~{S},8~{a}~{S})-3-[(4-fluoranyl-1~{H}-indol-3-yl)methyl]-2,3,6,7,8,8~{a}-hexahydropyrrolo[1,2-a]pyrazine-1,4-dione, Cytochrome P450-F5053, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ma, B.D, Tian, W, Qu, X, Kong, X.D. | | Deposit date: | 2022-12-09 | | Release date: | 2023-04-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineering the Substrate Specificity of a P450 Dimerase Enables the Collective Biosynthesis of Heterodimeric Tryptophan-Containing Diketopiperazines.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8HN0
 
 | |
5H49
 
 | | Crystal structure of Cbln1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cerebellin-1 | | Authors: | Zhong, C, Shen, J, Zhang, H, Ding, J. | | Deposit date: | 2016-10-31 | | Release date: | 2017-09-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cbln1 and Cbln4 Are Structurally Similar but Differ in GluD2 Binding Interactions.
Cell Rep, 20, 2017
|
|
3RO5
 
 | | Crystal structure of influenza A virus nucleoprotein with ligand | | Descriptor: | Nucleocapsid protein, [4-(2-chloro-4-nitrophenyl)piperazin-1-yl][3-(2-methoxyphenyl)-5-methyl-1,2-oxazol-4-yl]methanone | | Authors: | Pearce, B.C, Edavettal, S, McDonnell, P.A, Lewis, H.A, Steinbacher, S, Baldwin, E.T, Langley, D.R, Maskos, K, Mortl, M, Kiefersauer, R. | | Deposit date: | 2011-04-25 | | Release date: | 2011-09-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Inhibition of influenza virus replication via small molecules that induce the formation of higher-order nucleoprotein oligomers.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4P46
 
 | | J809.B5 Y31A TCR bound to IAb3K | | Descriptor: | 3K Peptide,H-2 class II histocompatibility antigen, A beta chain, H-2 class II histocompatibility antigen, ... | | Authors: | Stadinski, B.D, Huseby, E.S, Trenh, P, Stern, L.J. | | Deposit date: | 2014-03-11 | | Release date: | 2014-05-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.851 Å) | | Cite: | Effect of CDR3 Sequences and Distal V Gene Residues in Regulating TCR-MHC Contacts and Ligand Specificity.
J Immunol., 192, 2014
|
|
4P5T
 
 | | 14.C6 TCR complexed with MHC class II I-Ab/3K peptide | | Descriptor: | H-2 class II histocompatibility antigen, A-B alpha chain, Human nkt tcr beta chain, ... | | Authors: | Trenh, P, Stadinski, B, Huseby, E.S, Stern, L.J. | | Deposit date: | 2014-03-19 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.263 Å) | | Cite: | Effect of CDR3 Sequences and Distal V Gene Residues in Regulating TCR-MHC Contacts and Ligand Specificity.
J Immunol., 192, 2014
|
|
4P23
 
 | | J809.B5 TCR bound to IAb/3K | | Descriptor: | 3K peptide and MHC IAb beta chain,H-2 class II histocompatibility antigen, A beta chain, H-2 class II histocompatibility antigen, ... | | Authors: | Stadinski, B.D, Huseby, E.S, Trenh, P, Stern, L. | | Deposit date: | 2014-02-28 | | Release date: | 2014-05-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Effect of CDR3 Sequences and Distal V Gene Residues in Regulating TCR-MHC Contacts and Ligand Specificity.
J Immunol., 192, 2014
|
|
2L89
 
 | |
5ZNK
 
 | | Crystal structure of a bacterial ProRS with ligands | | Descriptor: | 7-chloro-6-fluoro-3-{2-oxo-3-[(2S)-piperidin-2-yl]propyl}quinazolin-4(3H)-one, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Cheng, B, Yu, Y, Zhou, H. | | Deposit date: | 2018-04-09 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Guided Design of Halofuginone Derivatives as ATP-Aided Inhibitors Against Bacterial Prolyl-tRNA Synthetase.
J.Med.Chem., 65, 2022
|
|
6BOJ
 
 | | Crystal Structure of the PDE4D Catalytic Domain and UCR2 Regulatory Helix with BPN5004 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(4-{[2-(3-chlorophenyl)-6-ethylpyrimidin-4-yl]methyl}phenyl)acetamide, CHLORIDE ION, ... | | Authors: | Fox III, D, Fairman, J.W, Gurney, M.E. | | Deposit date: | 2017-11-20 | | Release date: | 2018-08-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Memory enhancing effects of BPN14770, an allosteric inhibitor of phosphodiesterase-4D, in wild-type and humanized mice.
Neuropsychopharmacology, 43, 2018
|
|
5ZNJ
 
 | | Crystal structure of a bacterial ProRS with ligands | | Descriptor: | 7-bromo-6-chloro-3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Cheng, B, Yu, Y, Zhou, H. | | Deposit date: | 2018-04-09 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structure-Guided Design of Halofuginone Derivatives as ATP-Aided Inhibitors Against Bacterial Prolyl-tRNA Synthetase.
J.Med.Chem., 65, 2022
|
|
7KW1
 
 | | Structure of hSTING in complex with novel carbocyclic pyrimidine CDN-3 | | Descriptor: | (2R,5R,7R,8R,10R,12aR,14R,15aS,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-16-hydroxy-14-[(pyrimidin-4-yl)oxy]-2,10-disulfanyldecahydro-2H,10H-5,8-methano-2lambda~5~,10lambda~5~-cyclopenta[l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Skene, R. | | Deposit date: | 2020-11-29 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of Novel Carbocyclic Pyrimidine Cyclic Dinucleotide STING Agonists for Antitumor Immunotherapy Using Systemic Intravenous Route.
J.Med.Chem., 64, 2021
|
|
7KVX
 
 | | Structure of hSTING in complex with novel carbocyclic pyrimidine CDN 1 | | Descriptor: | (2R,5R,7R,8R,10R,12aR,14R,15aS,16R)-7-(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)-16-hydroxy-14-[(pyrimidin-4-yl)amino]-2,10-disulfanyldecahydro-2H,10H-5,8-methano-2lambda~5~,10lambda~5~-cyclopenta[l][1,3,6,9,11,2,10]pentaoxadiphosphacyclotetradecine-2,10-dione, Stimulator of interferon genes protein | | Authors: | Skene, R. | | Deposit date: | 2020-11-29 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Identification of Novel Carbocyclic Pyrimidine Cyclic Dinucleotide STING Agonists for Antitumor Immunotherapy Using Systemic Intravenous Route.
J.Med.Chem., 64, 2021
|
|
