7DFP
 
 | | Human dopamine D2 receptor in complex with spiperone | | Descriptor: | 8-[4-(4-fluorophenyl)-4-oxidanylidene-butyl]-1-phenyl-1,3,8-triazaspiro[4.5]decan-4-one, D(2) dopamine receptor,Soluble cytochrome b562, FabH, ... | | Authors: | Im, D, Shimamura, T, Iwata, S. | | Deposit date: | 2020-11-09 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the dopamine D 2 receptor in complex with the antipsychotic drug spiperone.
Nat Commun, 11, 2020
|
|
7VJ6
 
 | | SFX structure of archaeal class II CPD photolyase from Methanosarcina mazei in the semiquinone state | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.C, Nango, E, Gusti-Ngurah-Putu, E.-P, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Sugahara, M, Owada, S, Joti, Y, Tanaka, R, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJ7
 
 | | SFX structure of archaeal class II CPD photolyase from Methanosarcina mazei in the fully reduced state | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.C, Nango, E, Gusti-Ngurah-Putu, E.-P, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Sugahara, M, Owada, S, Joti, Y, Tanaka, R, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
7VJ8
 
 | | SFX structure of archaeal class II CPD photolyase from Methanosarcina mazei in the oxidized state | | Descriptor: | DNA photolyase, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Maestre-Reyna, M, Yang, C.-H, Huang, W.C, Nango, E, Gusti-Ngurah-Putu, E.-P, Franz-Badur, S, Wu, W.-J, Wu, H.-Y, Wang, P.-H, Liao, J.-H, Lee, C.-C, Huang, K.-F, Chang, Y.-K, Weng, J.-H, Sugahara, M, Owada, S, Joti, Y, Tanaka, R, Tono, K, Kiontke, S, Yamamoto, J, Iwata, S, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Serial crystallography captures dynamic control of sequential electron and proton transfer events in a flavoenzyme.
Nat.Chem., 14, 2022
|
|
5YJP
 
 | | Human chymase in complex with 3-(ethoxyimino)-7-oxo-1,4-diazepane derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-((R)-1-((R,Z)-6-(5-chloro-2-methoxybenzyl)-3-(ethoxyimino)-7-oxo-1,4-diazepane-1-carboxamido)propyl)benzoic acid, ZINC ION, ... | | Authors: | Sugawara, H. | | Deposit date: | 2017-10-11 | | Release date: | 2017-12-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design, synthesis, and binding mode analysis of novel and potent chymase inhibitors
Bioorg. Med. Chem. Lett., 28, 2018
|
|
5YC5
 
 | | Crystal structure of human IgG-Fc in complex with aglycan and optimized Fc gamma receptor IIIa | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Immunoglobulin gamma-1 heavy chain, ... | | Authors: | Caaveiro, J.M.M, Tamura, H, Tsumoto, K, Kiyoshi, M. | | Deposit date: | 2017-09-06 | | Release date: | 2018-03-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Assessing the Heterogeneity of the Fc-Glycan of a Therapeutic Antibody Using an engineered Fc gamma Receptor IIIa-Immobilized Column.
Sci Rep, 8, 2018
|
|
5YJM
 
 | | Human chymase in complex with 7-oxo-3-(phenoxyimino)-1,4-diazepane derivative | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-4-((R)-1-((R,Z)-6-(5-chloro-2-methoxybenzyl)-7-oxo-3-(phenoxyimino)-1,4-diazepane-1-carboxamido)propyl)benzoic acid, ZINC ION, ... | | Authors: | Sugawara, H. | | Deposit date: | 2017-10-11 | | Release date: | 2017-12-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based design, synthesis, and binding mode analysis of novel and potent chymase inhibitors
Bioorg. Med. Chem. Lett., 28, 2018
|
|
1DLF
 
 | | HIGH RESOLUTION CRYSTAL STRUCTURE OF THE FV FRAGMENT FROM AN ANTI-DANSYL SWITCH VARIANT ANTIBODY IGG2A(S) CRYSTALLIZED AT PH 5.25 | | Descriptor: | ANTI-DANSYL IMMUNOGLOBULIN IGG2A(S), SULFATE ION | | Authors: | Nakasako, M, Takahashi, H, Shimada, I, Arata, Y. | | Deposit date: | 1998-07-14 | | Release date: | 1999-07-26 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The pH-dependent structural variation of complementarity-determining region H3 in the crystal structures of the Fv fragment from an anti-dansyl monoclonal antibody.
J.Mol.Biol., 291, 1999
|
|
2D7E
 
 | | Crystal structure of N-terminal domain of PriA from E.coli | | Descriptor: | Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Maenaka, K, Masai, H, Kohda, D. | | Deposit date: | 2005-11-18 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2D7H
 
 | | Crystal structure of the ccc complex of the N-terminal domain of PriA | | Descriptor: | DNA (5'-D(P*CP*CP*C)-3'), Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Maenaka, K, Masai, H, Kohda, D. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
2D7G
 
 | | Crystal structure of the aa complex of the N-terminal domain of PriA | | Descriptor: | DNA (5'-D(P*AP*A)-3'), Primosomal protein N' | | Authors: | Sasaki, K, Ose, T, Maenaka, K, Masai, H, Kohda, D. | | Deposit date: | 2005-11-21 | | Release date: | 2006-11-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis of the 3'-end recognition of a leading strand in stalled replication forks by PriA.
EMBO J., 26, 2007
|
|
7E9L
 
 | | Crystal Structure of POMGNT2 in complex with UDP and mono-mannosyl peptide (379Man short peptide) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2021-03-04 | | Release date: | 2021-05-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of POMGNT2 provides new insights into the mechanism to determine the functional O-mannosylation site on alpha-dystroglycan.
Genes Cells, 26, 2021
|
|
7E9K
 
 | | Crystal Structure of POMGNT2 in complex with UDP and mono-mannosyl peptide (379Man long peptide) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2021-03-04 | | Release date: | 2021-05-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The structure of POMGNT2 provides new insights into the mechanism to determine the functional O-mannosylation site on alpha-dystroglycan.
Genes Cells, 26, 2021
|
|
7E9J
 
 | | Crystal Structure of POMGNT2 in complex with UDP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Protein O-linked-mannose beta-1,4-N-acetylglucosaminyltransferase 2, ... | | Authors: | Kuwabara, N. | | Deposit date: | 2021-03-04 | | Release date: | 2021-05-05 | | Last modified: | 2021-07-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The structure of POMGNT2 provides new insights into the mechanism to determine the functional O-mannosylation site on alpha-dystroglycan.
Genes Cells, 26, 2021
|
|
7BV9
 
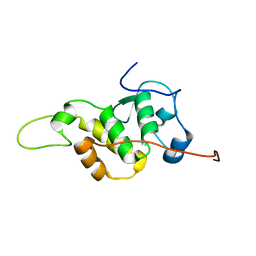 | | The NMR structure of the BEN domain from human NAC1 | | Descriptor: | Nucleus accumbens-associated protein 1 | | Authors: | Nagata, T, Kobayashi, N, Nakayama, N, Obayashi, E, Urano, T. | | Deposit date: | 2020-04-09 | | Release date: | 2021-02-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Nucleus Accumbens-Associated Protein 1 Binds DNA Directly through the BEN Domain in a Sequence-Specific Manner.
Biomedicines, 8, 2020
|
|
2EB3
 
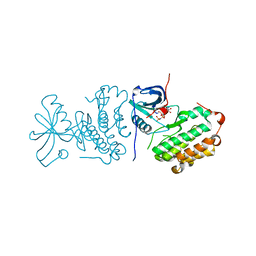 | | Crystal structure of mutated EGFR kinase domain (L858R) in complex with AMPPNP | | Descriptor: | Epidermal growth factor receptor, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Senba, K, Yamamoto, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-06 | | Release date: | 2008-02-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor
Oncogene, 2012
|
|
2EB2
 
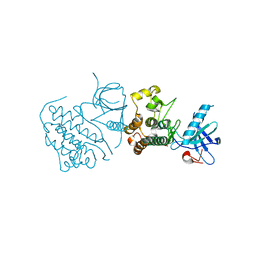 | | Crystal structure of mutated EGFR kinase domain (G719S) | | Descriptor: | Epidermal growth factor receptor | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Chen, L, Liu, Z.J, Wang, B.C, Shirouzu, M, Senba, K, Yamamoto, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-06 | | Release date: | 2008-02-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor
Oncogene, 2012
|
|
3UG2
 
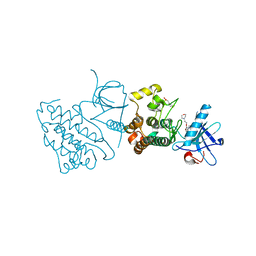 | | Crystal structure of the mutated EGFR kinase domain (G719S/T790M) in complex with gefitinib | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Epidermal growth factor receptor, Gefitinib | | Authors: | Parker, L.J, Handa, N, Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor
Oncogene, 32, 2013
|
|
3VJO
 
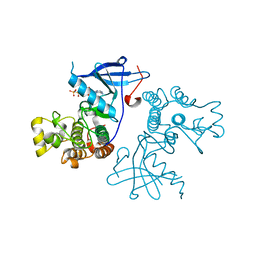 | | Crystal structure of the wild-type EGFR kinase domain in complex with AMPPNP. | | Descriptor: | Epidermal growth factor receptor, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Semba, K, Yamamoto, T, Yokoyama, S. | | Deposit date: | 2011-10-27 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor.
Oncogene, 32, 2013
|
|
3W32
 
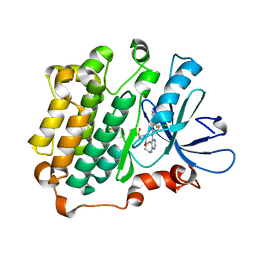 | | EGFR kinase domain complexed with compound 20a | | Descriptor: | 4-({3-chloro-4-[3-(trifluoromethyl)phenoxy]phenyl}amino)-N-[2-(methylsulfonyl)ethyl]-8,9-dihydro-7H-pyrimido[4,5-b]azepine-6-carboxamide, Epidermal growth factor receptor, SULFATE ION | | Authors: | Sogabe, S, Kawakita, Y. | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and synthesis of novel pyrimido[4,5-b]azepine derivatives as HER2/EGFR dual inhibitors
Bioorg.Med.Chem., 21, 2013
|
|
3UG1
 
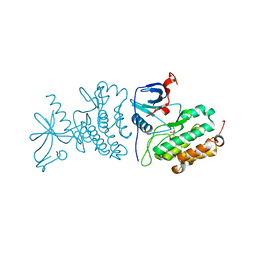 | | Crystal structure of the mutated EGFR kinase domain (G719S/T790M) in the apo form | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Epidermal growth factor receptor | | Authors: | Parker, L.J, Handa, N, Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Yokoyama, S. | | Deposit date: | 2011-11-02 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor
Oncogene, 32, 2013
|
|
3AQO
 
 | |
3W33
 
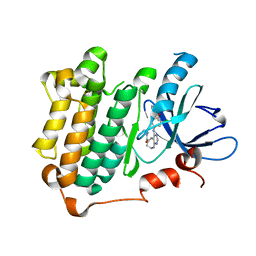 | | EGFR kinase domain complexed with compound 19b | | Descriptor: | 4-{[4-(1-benzothiophen-4-yloxy)-3-chlorophenyl]amino}-N-(2-hydroxyethyl)-8,9-dihydro-7H-pyrimido[4,5-b]azepine-6-carboxamide, Epidermal growth factor receptor, SULFATE ION | | Authors: | Sogabe, S, Kawakita, Y. | | Deposit date: | 2012-12-07 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Design and synthesis of novel pyrimido[4,5-b]azepine derivatives as HER2/EGFR dual inhibitors
Bioorg.Med.Chem., 21, 2013
|
|
3VJN
 
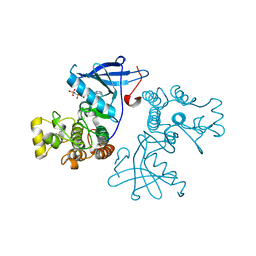 | | Crystal structure of the mutated EGFR kinase domain (G719S/T790M) in complex with AMPPNP. | | Descriptor: | Epidermal growth factor receptor, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Yoshikawa, S, Kukimoto-Niino, M, Shirouzu, M, Semba, K, Yamamoto, T, Yokoyama, S. | | Deposit date: | 2011-10-27 | | Release date: | 2012-03-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural basis for the altered drug sensitivities of non-small cell lung cancer-associated mutants of human epidermal growth factor receptor.
Oncogene, 32, 2013
|
|
2ZDU
 
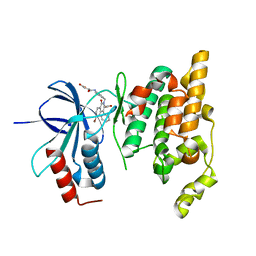 | | Crystal Structure of human JNK3 complexed with an isoquinolone inhibitor | | Descriptor: | 4-[(4-{[6-bromo-3-(methoxycarbonyl)-1-oxo-4-phenylisoquinolin-2(1H)-yl]methyl}phenyl)amino]-4-oxobutanoic acid, Mitogen-activated protein kinase 10 | | Authors: | Sogabe, S, Ohra, T, Itoh, F, Habuka, N, Fujishima, A. | | Deposit date: | 2007-11-27 | | Release date: | 2008-09-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery, synthesis and biological evaluation of isoquinolones as novel and highly selective JNK inhibitors (1)
Bioorg.Med.Chem., 16, 2008
|
|
