7CH9
 
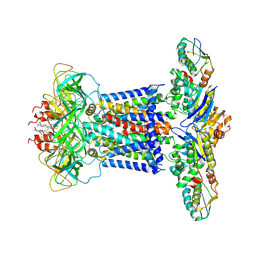 | | Cryo-EM structure of P.aeruginosa MlaFEBD | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, MlaD domain-containing protein, Probable ATP-binding component of ABC transporter, ... | | Authors: | Zhou, C, Shi, H, Zhang, M, Huang, Y. | | Deposit date: | 2020-07-05 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural Insight into Phospholipid Transport by the MlaFEBD Complex from P. aeruginosa.
J.Mol.Biol., 433, 2021
|
|
7ESI
 
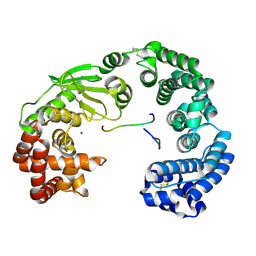 | | Crystal structure of the collagenase unit of a Vibrio collagenase from Vibrio harveyi VHJR7 at 1. 8 angstrom resolution. | | Descriptor: | CALCIUM ION, Collagenase unit (CU), Peptide P1, ... | | Authors: | Cao, H.Y, Wang, Y, Peng, M, Zhang, Y.Z. | | Deposit date: | 2021-05-11 | | Release date: | 2022-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of Vibrio collagenase VhaC provides insight into the mechanism of bacterial collagenolysis.
Nat Commun, 13, 2022
|
|
7EV3
 
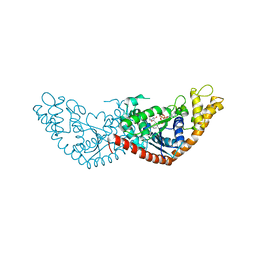 | |
7EV2
 
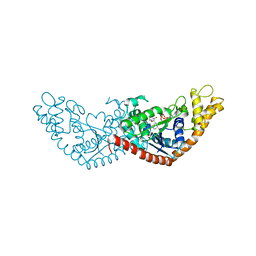 | |
7YVC
 
 | | Aplysia californica FaNaC in apo state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Chen, Q.F, Liu, F.L, Dang, Y, Feng, H, Zhang, Z, Ye, S. | | Deposit date: | 2022-08-19 | | Release date: | 2023-08-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and mechanism of a neuropeptide-activated channel in the ENaC/DEG superfamily.
Nat.Chem.Biol., 19, 2023
|
|
7YVB
 
 | | Aplysia californica FaNaC in ligand bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FMRFamide-gated Na+ channel, ... | | Authors: | Chen, Q.F, Liu, F.L, Dang, Y, Feng, H, Zhang, Z, Ye, S. | | Deposit date: | 2022-08-19 | | Release date: | 2023-08-09 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and mechanism of a neuropeptide-activated channel in the ENaC/DEG superfamily.
Nat.Chem.Biol., 19, 2023
|
|
7CRC
 
 | | Cryo-EM structure of plant NLR RPP1 tetramer in complex with ATR1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Avirulence protein ATR1, ... | | Authors: | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | Deposit date: | 2020-08-13 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
6KUU
 
 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B3) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-12-11 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B3)
To Be Published
|
|
7CRB
 
 | | Cryo-EM structure of plant NLR RPP1 LRR-ID domain in complex with ATR1 | | Descriptor: | Avirulence protein ATR1, NAD+ hydrolase (NADase) | | Authors: | Ma, S.C, Lapin, D, Liu, L, Sun, Y, Song, W, Zhang, X.X, Logemann, E, Yu, D.L, Wang, J, Jirschitzka, J, Han, Z.F, SchulzeLefert, P, Parker, J.E, Chai, J.J. | | Deposit date: | 2020-08-13 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme.
Science, 370, 2020
|
|
8INA
 
 | | Crystal structure of UGT74AN3-UDP | | Descriptor: | GLYCEROL, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE | | Authors: | Long, F, Huang, W. | | Deposit date: | 2023-03-09 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8IND
 
 | | Crystal structure of UGT74AN3-UDP-RES | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5-[(1R,2S,4R,6R,7R,10S,11S,14S,16R)-14-hydroxy-7,11-dimethyl-3-oxapentacyclo[8.8.0.02,4.02,7.011,16]octadecan-6-yl]pyran-2-one, Glycosyltransferase, ... | | Authors: | Huang, W. | | Deposit date: | 2023-03-09 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8INV
 
 | | Crystal structure of UGT74AN3-UDP-BUF | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycosyltransferase, URIDINE-5'-DIPHOSPHATE, ... | | Authors: | Long, F, Huang, W. | | Deposit date: | 2023-03-10 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8WGX
 
 | | Cryo-EM structure of inward-open state human norepinephrine transporter NET bound with norepinephrine in nanodisc. | | Descriptor: | L-NOREPINEPHRINE, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2023-09-22 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 632, 2024
|
|
8INO
 
 | | Crystal structure of UGT74AN3 in complex UDP and PER | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-[(3S,5S,8S,9S,10R,13R,14S,17R)-10,13-dimethyl-3,5,14-tris(oxidanyl)-2,3,4,6,7,8,9,11,12,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthren-17-yl]-2H-furan-5-one, Glycosyltransferase, ... | | Authors: | Long, F, Huang, W. | | Deposit date: | 2023-03-10 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Substrate Promiscuity, Crystal Structure, and Application of a Plant UDP-Glycosyltransferase UGT74AN3
Acs Catalysis, 14, 2024
|
|
8WGR
 
 | | Cryo-EM structure of inward-open state human norepinephrine transporter NET bound with antidepressant desipramine in KCl condition. | | Descriptor: | 3-(10,11-DIHYDRO-5H-DIBENZO[B,F]AZEPIN-5-YL)-N-METHYLPROPAN-1-AMINE, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2023-09-22 | | Release date: | 2024-07-31 | | Last modified: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 632, 2024
|
|
8YY8
 
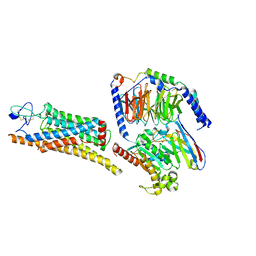 | | Fzd7 -Gs complex | | Descriptor: | Frizzled-7, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Chen, B, Xu, L, Han, G.W, Xu, F. | | Deposit date: | 2024-04-03 | | Release date: | 2024-04-24 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Cryo-EM structure of constitutively active human Frizzled 7 in complex with heterotrimeric G s .
Cell Res., 31, 2021
|
|
8K4H
 
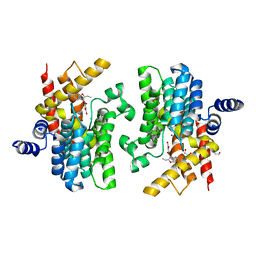 | | Crystal structure of PDE4D complexed with benzbromarone | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, ZINC ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-07-18 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
8K4C
 
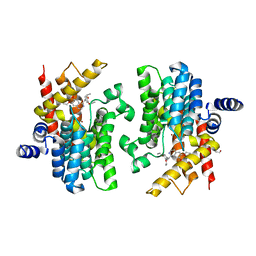 | | Crystal structure of PDE4D complexed with ethaverine hydrochloride | | Descriptor: | 1,2-ETHANEDIOL, 1-[(3,4-diethoxyphenyl)methyl]-6,7-diethoxy-isoquinoline, MAGNESIUM ION, ... | | Authors: | Liu, J.Y, Li, M.J, Xu, Y.C. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Drug repurposing and structure-based discovery of new PDE4 and PDE5 inhibitors.
Eur.J.Med.Chem., 262, 2023
|
|
8K71
 
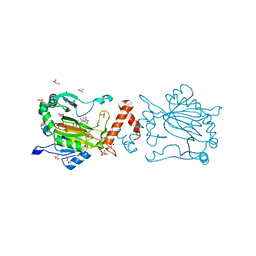 | | Factor-inhibiting hypoxia-inducible factor in complex with Zn(II) and 2-(3-hydroxy-2-((2-((naphthalen-2-ylmethyl)sulfonyl)acetyl)imino)-2,3-dihydrothiazol-4-yl)acetic acid | | Descriptor: | 2-[(2~{Z})-2-[2-(naphthalen-2-ylmethylsulfonyl)ethanoylimino]-3-oxidanyl-1,3-thiazol-4-yl]ethanoic acid, GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Nakashima, Y, Corner, T.P, Teo, R.Z.R, Brewitz, L, Schofield, C.J. | | Deposit date: | 2023-07-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structure-guided optimisation of N -hydroxythiazole-derived inhibitors of factor inhibiting hypoxia-inducible factor-alpha.
Chem Sci, 14, 2023
|
|
8K72
 
 | | Factor-inhibiting hypoxia-inducible factor in complex with Zn(II) and 2-(3-hydroxy-2-((3-(phenylsulfonamido)propanoyl)imino)-2,3-dihydrothiazol-4-yl)acetic acid | | Descriptor: | 2-[(2~{Z})-3-oxidanyl-2-[3-(phenylsulfonylamino)propanoylimino]-1,3-thiazol-4-yl]ethanoic acid, GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Nakashima, Y, Corner, T.P, Teo, R.Z.R, Brewitz, L, Schofield, C.J. | | Deposit date: | 2023-07-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure-guided optimisation of N -hydroxythiazole-derived inhibitors of factor inhibiting hypoxia-inducible factor-alpha.
Chem Sci, 14, 2023
|
|
8K73
 
 | | Factor-inhibiting hypoxia-inducible factor in complex with Zn(II) and 2-(3-hydroxy-2-((1-(phenylsulfonyl)pyrrolidine-3-carbonyl)imino)-2,3-dihydrothiazol-4-yl)acetic acid | | Descriptor: | 2-[(2~{Z})-3-oxidanyl-2-[(3~{R})-1-(phenylsulfonyl)pyrrolidin-3-yl]carbonylimino-1,3-thiazol-4-yl]ethanoic acid, GLYCEROL, Hypoxia-inducible factor 1-alpha inhibitor, ... | | Authors: | Nakashima, Y, Corner, T.P, Teo, R.Z.R, Brewitz, L, Schofield, C.J. | | Deposit date: | 2023-07-26 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure-guided optimisation of N -hydroxythiazole-derived inhibitors of factor inhibiting hypoxia-inducible factor-alpha.
Chem Sci, 14, 2023
|
|
7YED
 
 | | In situ structure of polymerase complex of mammalian reovirus in the elongation state | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Lambda-2 protein, MAGNESIUM ION, ... | | Authors: | Bao, K.Y, Zhang, X.L, Li, D.Y, Zhu, P. | | Deposit date: | 2022-07-05 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | In situ structures of polymerase complex of mammalian reovirus illuminate RdRp activation and transcription regulation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XUZ
 
 | | Crystal structure of a HDAC4-MEF2A-DNA ternary complex | | Descriptor: | DNA (5'-D(*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*T)-3'), DNA (5'-D(P*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*A)-3'), Histone deacetylase 4, ... | | Authors: | Dai, S.Y, Guo, L, Dey, R, Guo, M, Bates, D, Cayford, J, Chen, X.J, Wei, X.D, Chen, L, Chen, Y.H. | | Deposit date: | 2022-05-20 | | Release date: | 2023-11-29 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (3.591 Å) | | Cite: | Structural insights into the HDAC4-MEF2A-DNA complex and its implication in long-range transcriptional regulation.
Nucleic Acids Res., 52, 2024
|
|
8Z1L
 
 | | Cryo-EM structure of human norepinephrine transporter NET in the presence of the antidepressant atomoxetine in an outward-open state at resolution of 3.4 angstrom. | | Descriptor: | (3R)-N-methyl-3-(2-methylphenoxy)-3-phenyl-propan-1-amine, CHLORIDE ION, Sodium-dependent noradrenaline transporter | | Authors: | Tan, J, Xiao, Y, Kong, F, Lei, J, Yuan, Y, Yan, C. | | Deposit date: | 2024-04-11 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular basis of human noradrenaline transporter reuptake and inhibition.
Nature, 632, 2024
|
|
7YEZ
 
 | | In situ structure of polymerase complex of mammalian reovirus in the reloaded state | | Descriptor: | Lambda-2 protein, Mu-2 protein, RNA helicase, ... | | Authors: | Bao, K.Y, Zhang, X.L, Li, D.Y, Zhu, P. | | Deposit date: | 2022-07-06 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | In situ structures of polymerase complex of mammalian reovirus illuminate RdRp activation and transcription regulation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
