2I1N
 
 | | Crystal structure of the 1st PDZ domain of Human DLG3 | | Descriptor: | Discs, large homolog 3, SODIUM ION | | Authors: | Turnbull, A.P, Phillips, C, Bunkoczi, G, Debreczeni, J, Ugochukwu, E, Pike, A.C.W, Gorrec, F, Umeano, C, Elkins, J, Berridge, G, Savitsky, P, Gileadi, O, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-08-14 | | Release date: | 2006-09-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions.
Protein Sci., 16, 2007
|
|
2JII
 
 | | Structure of vaccinia related kinase 3 | | Descriptor: | 1,2-ETHANEDIOL, SERINE/THREONINE-PROTEIN KINASE VRK3 MOLECULE: VACCINIA RELATED KINASE 3 | | Authors: | Bunkoczi, G, Eswaran, J, Pike, A.C.W, Uppenberg, J, Ugochukwu, E, von Delft, F, Cooper, C, Salah, E, Savitsky, P, Burgess-Brown, N, Keates, T, Fedorov, O, Sobott, F, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Weigelt, J, Knapp, S. | | Deposit date: | 2007-06-28 | | Release date: | 2007-07-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the pseudokinase VRK3 reveals a degraded catalytic site, a highly conserved kinase fold, and a putative regulatory binding site.
Structure, 17, 2009
|
|
6Y5V
 
 | | Structure of Human Potassium Chloride Transporter KCC3b (S45D/T940D/T997D) in KCl | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Solute carrier family 12 member 6, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Chi, G, Man, H, Ebenhoch, R, Reggiano, G, Pike, A.C.W, Wang, D, McKinley, G, Mukhopadhyay, S.M.M, MacLean, E.M, Chalk, R, Moreau, C, Snee, M, Bohstedt, T, Singh, N.K, Abrusci, P, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Marsden, B.D, Burgess-Brown, N.A, DiMaio, F, Duerr, K.L. | | Deposit date: | 2020-02-25 | | Release date: | 2020-07-15 | | Last modified: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Phospho-regulation, nucleotide binding and ion access control in potassium-chloride cotransporters.
Embo J., 40, 2021
|
|
4AP2
 
 | | Crystal structure of the human KLHL11-Cul3 complex at 2.8A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CULLIN-3, ... | | Authors: | Canning, P, Cooper, C.D.O, Krojer, T, Filippakopoulos, P, Ayinampudi, V, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-03-30 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for Cul3 Assembly with the Btb-Kelch Family of E3 Ubiquitin Ligases.
J.Biol.Chem., 288, 2013
|
|
4ASC
 
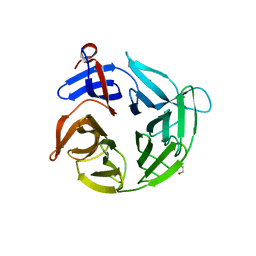 | | Crystal structure of the Kelch domain of human KBTBD5 | | Descriptor: | 1,2-ETHANEDIOL, KELCH REPEAT AND BTB DOMAIN-CONTAINING PROTEIN 5 | | Authors: | Canning, P, Ayinampudi, V, Krojer, T, Strain-Damerell, C, Raynor, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structural Basis for Cul3 Assembly with the Btb-Kelch Family of E3 Ubiquitin Ligases.
J.Biol.Chem., 288, 2013
|
|
4APF
 
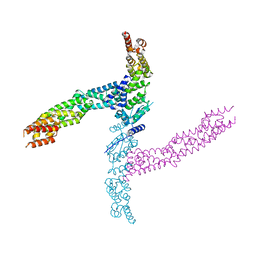 | | Crystal structure of the human KLHL11-Cul3 complex at 3.1A resolution | | Descriptor: | CULLIN 3, KELCH-LIKE PROTEIN 11 | | Authors: | Canning, P, Cooper, C.D.O, Krojer, T, Vollmar, M, Ugochukwu, E, Muniz, J.R.C, Ayinampudi, V, Savitsky, P, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F, Bullock, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-04-02 | | Release date: | 2012-05-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Basis for Cul3 Assembly with the Btb-Kelch Family of E3 Ubiquitin Ligases.
J.Biol.Chem., 288, 2013
|
|
3H9R
 
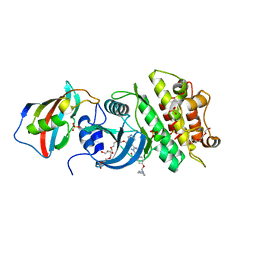 | | Crystal structure of the kinase domain of type I activin receptor (ACVR1) in complex with FKBP12 and dorsomorphin | | Descriptor: | 6-[4-(2-piperidin-1-ylethoxy)phenyl]-3-pyridin-4-ylpyrazolo[1,5-a]pyrimidine, Activin receptor type-1, Peptidyl-prolyl cis-trans isomerase FKBP1A, ... | | Authors: | Chaikuad, A, Alfano, I, Shrestha, B, Muniz, J.R.C, Petrie, K, Fedorov, O, Phillips, C, Bishop, S, Mahajan, P, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-04-30 | | Release date: | 2009-06-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure of the Bone Morphogenetic Protein Receptor ALK2 and Implications for Fibrodysplasia Ossificans Progressiva.
J.Biol.Chem., 287, 2012
|
|
3GVU
 
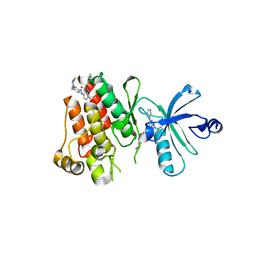 | | The crystal structure of human ABL2 in complex with GLEEVEC | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, Tyrosine-protein kinase ABL2 | | Authors: | Ugochukwu, E, Salah, E, Barr, A, Mahajan, P, Shrestha, B, Savitsky, P, Chaikuad, A, Filippakopoulos, P, Roos, A, Pike, A.C.W, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-03-31 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The crystal structure of human ABL2 in complex with GLEEVEC
To be Published
|
|
3IQ7
 
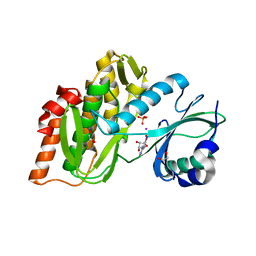 | | Crystal Structure of human Haspin in complex with 5-Iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 1,2-ETHANEDIOL, IODIDE ION, ... | | Authors: | Filippakopoulos, P, Eswaran, J, Keates, T, Burgess-Brown, N, Fedorov, O, Pike, A.C.W, Von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-19 | | Release date: | 2009-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and functional characterization of the atypical human kinase haspin.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2W2I
 
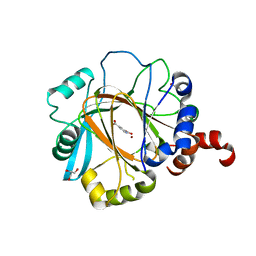 | | Crystal structure of the human 2-oxoglutarate oxygenase LOC390245 | | Descriptor: | 2-OXOGLUTARATE OXYGENASE, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Yue, W.W, Ng, S, Shafqat, N, Ugochukwu, E, McDonough, M, Pike, A.C.W, Filippakopoulos, P, von Delft, F, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Schofield, C, Oppermann, U. | | Deposit date: | 2008-10-31 | | Release date: | 2009-01-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Evolutionary Basis for the Dual Substrate Selectivity of Human Kdm4 Histone Demethylase Family.
J.Biol.Chem., 286, 2011
|
|
6FDZ
 
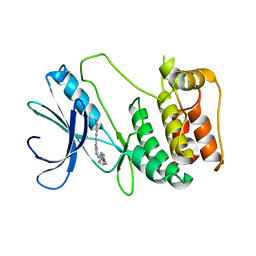 | | Unc-51-Like Kinase 3 (ULK3) In Complex With Momelotinib | | Descriptor: | Momelotinib, Serine/threonine-protein kinase ULK3 | | Authors: | Mathea, S, Salah, E, Moroglu, M, Scorah, A, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Huber, K, Knapp, S. | | Deposit date: | 2017-12-27 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Unc-51-Like Kinase 3 (ULK3) In Complex With Momelotinib
To Be Published
|
|
6FT3
 
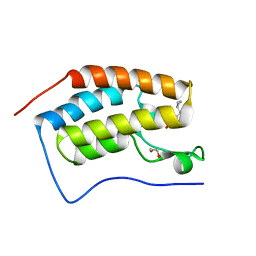 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-[(~{R})-cyclopropyl(oxidanyl)methyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)phenol, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Pike, A.C.W, Krojer, T, Conway, S.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-20 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | BET bromodomain ligands: Probing the WPF shelf to improve BRD4 bromodomain affinity and metabolic stability.
Bioorg.Med.Chem., 26, 2018
|
|
3K2L
 
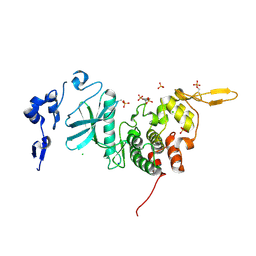 | | Crystal Structure of dual-specificity tyrosine phosphorylation regulated kinase 2 (DYRK2) | | Descriptor: | CHLORIDE ION, Dual specificity tyrosine-phosphorylation-regulated kinase 2, SODIUM ION, ... | | Authors: | Filippakopoulos, P, Myrianthopoulos, V, Soundararajan, M, Krojer, T, Hapka, E, Fedorov, O, Berridge, G, Wang, J, Shrestha, L, Pike, A.C.W, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Mikros, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structures of Down Syndrome Kinases, DYRKs, Reveal Mechanisms of Kinase Activation and Substrate Recognition.
Structure, 21, 2013
|
|
2WA0
 
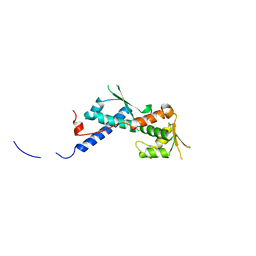 | | Crystal structure of the human MAGEA4 | | Descriptor: | MELANOMA-ASSOCIATED ANTIGEN 4 | | Authors: | Roos, A.K, Cooper, C.D.O, Ugochukwu, E, W Yue, W, Berridge, G, Elkins, J.M, Pike, A.C.W, Bray, J, Filippakopoulos, P, Muniz, J, Chaikuad, A, Burgess-Brown, N, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, von Delft, F, Gileadi, O, Oppermann, U. | | Deposit date: | 2009-01-31 | | Release date: | 2009-03-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of Two Melanoma-Associated Antigens Suggest Allosteric Regulation of Effector Binding.
Plos One, 11, 2016
|
|
2W2J
 
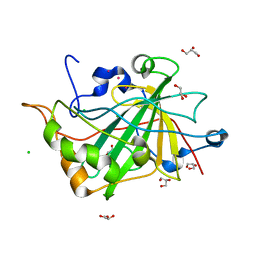 | | Crystal structure of the human carbonic anhydrase related protein VIII | | Descriptor: | CARBONIC ANHYDRASE-RELATED PROTEIN, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kramm, A, Muniz, J.R.C, Picaud, S.S, von Delft, F, Pilka, E.S, Yue, W.W, King, O.N.F, Kochan, G, Pike, A.C.W, Filippakopoulos, P, Arrowsmith, C, Wikstrom, M, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2008-10-31 | | Release date: | 2009-02-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of Human Carbonic Anhydrase-Related Protein Viii Reveals the Basis for Catalytic Silencing.
Proteins, 76, 2009
|
|
2WEI
 
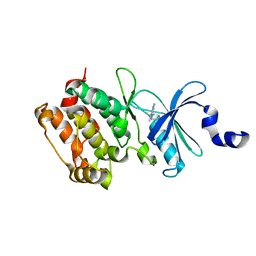 | | Crystal structure of the kinase domain of Cryptosporidium parvum calcium dependent protein kinase in complex with 3-MB-PP1 | | Descriptor: | 1-tert-butyl-3-(3-methylbenzyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, CALMODULIN-DOMAIN PROTEIN KINASE 1, PUTATIVE | | Authors: | Roos, A.K, King, O, Chaikuad, A, Zhang, C, Shokat, K.M, Wernimont, A.K, Artz, J.D, Lin, L, MacKenzie, F.I, Finerty, P.J, Vedadi, M, Schapira, M, Indarte, M, Kozieradzki, I, Pike, A.C.W, Fedorov, O, Doyle, D, Muniz, J, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, von Delft, F, Heightman, T, Hui, R. | | Deposit date: | 2009-03-31 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Cryptosporidium Parvum Kinome.
Bmc Genomics, 12, 2011
|
|
2VX3
 
 | | Crystal structure of the human dual specificity tyrosine- phosphorylation-regulated kinase 1A | | Descriptor: | CHLORIDE ION, DUAL SPECIFICITY TYROSINE-PHOSPHORYLATION- REGULATED KINASE 1A, HEXAETHYLENE GLYCOL, ... | | Authors: | Roos, A.K, Soundararajan, M, Pike, A.C.W, Federov, O, King, O, Burgess-Brown, N, Philips, C, Filippakopoulos, P, Arrowsmith, C.H, Wikstrom, M, Edwards, A, von Delft, F, Bountra, C, Knapp, S. | | Deposit date: | 2008-06-30 | | Release date: | 2008-09-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Down Syndrome Kinases, Dyrks, Reveal Mechanisms of Kinase Activation and Substrate Recognition.
Structure, 21, 2013
|
|
2V24
 
 | | Structure of the human SPRY domain-containing SOCS box protein SSB-4 | | Descriptor: | NICKEL (II) ION, SPRY DOMAIN-CONTAINING SOCS BOX PROTEIN 4 | | Authors: | Uppenberg, J, Bullock, A, Keates, T, Savitsky, P, Pike, A.C.W, Ugochukwu, E, Bunkoczi, G, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Knapp, S. | | Deposit date: | 2007-05-31 | | Release date: | 2007-07-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis for Par-4 Recognition by the Spry Domain-and Socs Box-Containing Proteins Spsb1, Spsb2, and Spsb4.
J.Mol.Biol., 401, 2010
|
|
2V4Z
 
 | | The crystal structure of the human G-protein subunit alpha (GNAI3) in complex with an engineered regulator of G-protein signaling type 2 domain (RGS2) | | Descriptor: | GUANINE NUCLEOTIDE-BINDING PROTEIN G(K) SUBUNIT ALPHA, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Roos, A.K, Soundararajan, M, Pike, A.C.W, Arrowsmith, C.H, Weigelt, J, Edwards, A, Bountra, C, Knapp, S. | | Deposit date: | 2008-09-30 | | Release date: | 2008-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Determinants of G-Protein Alpha Subunit Selectivity by Regulator of G-Protein Signaling 2(Rgs2).
J.Biol.Chem., 284, 2009
|
|
3K1Z
 
 | | Crystal Structure of Human Haloacid Dehalogenase-like Hydrolase Domain containing 3 (HDHD3) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Haloacid dehalogenase-like hydrolase domain-containing protein 3 | | Authors: | Ugochukwu, E, Guo, K, Yue, W.W, Pilka, E, Picaud, S, Muniz, J, Pike, A.C.W, Krojer, T, Gomes, M, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Kavanagh, K, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-29 | | Release date: | 2009-11-03 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Human Haloacid Dehalogenase-like Hydrolase Domain containing 3 (HDHD3)
To be Published
|
|
3K2O
 
 | | Structure of an oxygenase | | Descriptor: | ACETATE ION, Bifunctional arginine demethylase and lysyl-hydroxylase JMJD6, CHLORIDE ION, ... | | Authors: | Krojer, T, McDonough, M.A, Clifton, I.J, Mantri, M, Ng, S.S, Pike, A.C.W, Butler, D.S, Webby, C.J, Kochan, G, Bhatia, C, Bray, J.E, Chaikuad, A, Gileadi, O, von Delft, F, Weigelt, J, Arrowsmith, C.H, Bountra, C, Edwards, A.M, Schofield, C.J, Kavanagh, K.L, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-30 | | Release date: | 2009-11-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the 2-Oxoglutarate- and Fe(II)-Dependent Lysyl Hydroxylase JMJD6.
J.Mol.Biol., 401, 2010
|
|
2UZC
 
 | | Structure of human PDLIM5 in complex with the C-terminal peptide of human alpha-actinin-1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PDZ AND LIM DOMAIN 5 | | Authors: | Bunkoczi, G, Elkins, J, Salah, E, Burgess-Brown, N, Papagrigoriou, E, Pike, A.C.W, Turnbull, A, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A, Sundstrom, M, Weigelt, J, Doyle, D. | | Deposit date: | 2007-04-27 | | Release date: | 2007-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Unusual binding interactions in PDZ domain crystal structures help explain binding mechanisms.
Protein Sci., 19, 2010
|
|
2V1W
 
 | | Crystal structure of human LIM protein RIL (PDLIM4) PDZ domain bound to the C-terminal peptide of human alpha-actinin-1 | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, PDZ AND LIM DOMAIN PROTEIN 4, ... | | Authors: | Soundararajan, M, Shrestha, L, Pike, A.C.W, Salah, E, Burgess-Brown, N, Elkins, J, Umeano, C, Ugochukwu, E, von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Sundstrom, M, Doyle, D. | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Unusual Binding Interactions in Pdz Domain Crystal Structures Help Explain Binding Mechanisms.
Protein Sci., 19, 2010
|
|
6FSY
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | Descriptor: | 1,2-ETHANEDIOL, 3-(3,5-dimethyl-1,2-oxazol-4-yl)-5-[(~{R})-oxidanyl(pyridin-3-yl)methyl]phenol, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Conway, S.J, Pike, A.C.W, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-02-20 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | BET bromodomain ligands: Probing the WPF shelf to improve BRD4 bromodomain affinity and metabolic stability.
Bioorg.Med.Chem., 26, 2018
|
|
6FT4
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with a 3,5-dimethylisoxazol ligand | | Descriptor: | 3-[[4,4-bis(fluoranyl)piperidin-1-yl]methyl]-5-(3,5-dimethyl-1,2-oxazol-4-yl)phenol, Bromodomain-containing protein 4 | | Authors: | Filippakopoulos, P, Picaud, S, Pike, A.C.W, Krojer, T, Conway, S.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C. | | Deposit date: | 2018-02-20 | | Release date: | 2018-04-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | BET bromodomain ligands: Probing the WPF shelf to improve BRD4 bromodomain affinity and metabolic stability.
Bioorg.Med.Chem., 26, 2018
|
|
