6FXQ
 
 | | Structure of coproheme decarboxylase from Listeria monocytogenes during turnover | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, Putative heme-dependent peroxidase lmo2113, ... | | Authors: | Hofbauer, S, Pfanzagl, V, Mlynek, G, Puehringer, D. | | Deposit date: | 2018-03-09 | | Release date: | 2019-07-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Redox Cofactor Rotates during Its Stepwise Decarboxylation: Molecular Mechanism of Conversion of Coproheme to Hemeb.
Acs Catalysis, 9, 2019
|
|
4ZNN
 
 | | MicroED structure of the segment, GVVHGVTTVA, from the A53T familial mutant of Parkinson's disease protein, alpha-synuclein residues 47-56 | | Descriptor: | Alpha-synuclein | | Authors: | Rodriguez, J.A, Ivanova, M, Sawaya, M.R, Cascio, D, Reyes, F, Shi, D, Johnson, L, Guenther, E, Sangwan, S, Hattne, J, Nannenga, B, Brewster, A.S, Messerschmidt, M, Boutet, S, Sauter, N.K, Gonen, T, Eisenberg, D.S. | | Deposit date: | 2015-05-05 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-06 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.41 Å) | | Cite: | Structure of the toxic core of alpha-synuclein from invisible crystals.
Nature, 525, 2015
|
|
2XTL
 
 | | Structure of the major pilus backbone protein from Streptococcus Agalactiae | | Descriptor: | CELL WALL SURFACE ANCHOR FAMILY PROTEIN, POTASSIUM ION | | Authors: | Rinauda, D, Gourlay, L.J, Soriano, M, Grandi, G, Bolognesi, M. | | Deposit date: | 2010-10-11 | | Release date: | 2011-07-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure-Based Approach to Rationally Design a Chimeric Protein for an Effective Vaccine Against Group B Streptococcus Infections.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8FVT
 
 | |
8C7Z
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2308 | | Descriptor: | 1,2-ETHANEDIOL, 9-piperazin-1-yl-4-(3,4,5-trimethoxyphenyl)-5,6-dihydro-[1]benzoxepino[5,4-c]pyridine, AMMONIUM ION, ... | | Authors: | Cros, J, Williams, E.P, Sweeney, M.N, Smil, D, Gonzalez-Alvarez, H, Al-awar, R, Bullock, A.N. | | Deposit date: | 2023-01-18 | | Release date: | 2023-02-01 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Discovery of Conformationally Constrained ALK2 Inhibitors.
J.Med.Chem., 67, 2024
|
|
8C7W
 
 | | Crystal structure of the ACVR1 (ALK2) kinase in complex with the compound M4K2304 | | Descriptor: | 1,2-ETHANEDIOL, 6-methyl-9-piperazin-1-yl-4-(3,4,5-trimethoxyphenyl)-5,7-dihydropyrido[4,3-d][2]benzazepine, Activin receptor type I, ... | | Authors: | Cros, J, Williams, E.P, Sweeney, M.N, Smil, D, Gonzalez-Alvarez, H, Al-awar, R, Bullock, A.N. | | Deposit date: | 2023-01-17 | | Release date: | 2023-02-08 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of Conformationally Constrained ALK2 Inhibitors.
J.Med.Chem., 67, 2024
|
|
1YRA
 
 | | PAB0955 crystal structure : a GTPase in GDP bound form from Pyrococcus abyssi | | Descriptor: | ATP(GTP)binding protein, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Gras, S, Carpentier, P, Armengaud, J, Housset, D. | | Deposit date: | 2005-02-03 | | Release date: | 2006-02-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into a new homodimeric self-activated GTPase family.
Embo Rep., 8, 2007
|
|
2W9N
 
 | | crystal structure of linear di-ubiquitin | | Descriptor: | CHLORIDE ION, UBIQUITIN, ZINC ION | | Authors: | Komander, D, Reyes-Turcu, F, Wilkinson, K.D, Barford, D. | | Deposit date: | 2009-01-27 | | Release date: | 2009-04-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Molecular Discrimination of Structurally Equivalent Lys 63-Linked and Linear Polyubiquitin Chains.
Embo Rep., 10, 2009
|
|
2ZY0
 
 | | Crystal structure of the human RXR alpha ligand binding domain bound to a synthetic agonist compound and a coactivator peptide | | Descriptor: | 4-[2-(1,1,3,3-tetramethyl-2,3-dihydro-1H-1,3-benzodisilol-5-yl)-1,3-dioxolan-2-yl]benzoic acid, GRIP1 from Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Sato, Y, Antony, P, Rochel, N, Moras, D, Structural Genomics Consortium for Research on Gene Expression (SGCGES) | | Deposit date: | 2009-01-09 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Silicon analogues of the RXR-selective retinoid agonist SR11237 (BMS649): chemistry and biology
Chemmedchem, 4, 2009
|
|
2ZXZ
 
 | | Crystal structure of the human RXR alpha ligand binding domain bound to a synthetic agonist compound and a coactivator peptide | | Descriptor: | 4-[2-(1,1,3,3-tetramethyl-2,3-dihydro-1H-inden-5-yl)-1,3-dioxolan-2-yl]benzoic acid, GRIP1 from Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Sato, Y, Antony, P, Rochel, N, Moras, D, Structural Genomics Consortium for Research on Gene Expression (SGCGES) | | Deposit date: | 2009-01-09 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Silicon analogues of the RXR-selective retinoid agonist SR11237 (BMS649): chemistry and biology
Chemmedchem, 4, 2009
|
|
6MH2
 
 | | Structure of Herceptin Fab without antigen | | Descriptor: | Herceptin Fab arm heavy chain, Herceptin Fab arm light chain | | Authors: | Luthra, A, Langley, D.B, Christie, M, Christ, D. | | Deposit date: | 2018-09-17 | | Release date: | 2019-05-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human Antibody Bispecifics through Phage Display Selection.
Biochemistry, 58, 2019
|
|
8C3V
 
 | | SARS-CoV-2 Delta-RBD complexed with BA.2-13 Fab and C1 nanobody | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, BA.2-13 heavy chain, BA.2-13 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-12-28 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Rapid escape of new SARS-CoV-2 Omicron variants from BA.2-directed antibody responses.
Cell Rep, 42, 2023
|
|
1DNK
 
 | | THE X-RAY STRUCTURE OF THE DNASE I-D(GGTATACC)2 COMPLEX AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-D(*GP*GP*TP*AP*TP*AP*C)-3'), DNA (5'-D(*GP*GP*TP*AP*TP*AP*CP*C)-3'), ... | | Authors: | Weston, S.A, Lahm, A, Suck, D. | | Deposit date: | 1992-08-10 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray structure of the DNase I-d(GGTATACC)2 complex at 2.3 A resolution.
J.Mol.Biol., 226, 1992
|
|
6FTQ
 
 | |
2EJN
 
 | | Structural characterization of the tetrameric form of the major cat allergen fel D 1 | | Descriptor: | CALCIUM ION, Major allergen I polypeptide chain 1, chain 2 | | Authors: | Kaiser, L, Velickovic, T.C, Badia-Martinez, D, Adedoyin, J, Thunberg, S, Hallen, D, Berndt, K, Gronlund, H, Gafvelin, G, van Hage, M, Achour, A. | | Deposit date: | 2007-03-19 | | Release date: | 2007-04-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural characterization of the tetrameric form of the major cat allergen Fel d 1
J.Mol.Biol., 370, 2007
|
|
1ZTD
 
 | | Hypothetical Protein Pfu-631545-001 From Pyrococcus furiosus | | Descriptor: | Hypothetical Protein Pfu-631545-001 | | Authors: | Fu, Z.-Q, Horanyi, P, Florence, Q, Liu, Z.-J, Chen, L, Lee, D, Habel, J, Xu, H, Nguyen, D, Chang, S.-H, Zhou, W, Zhang, H, Jenney Jr, F.E, Sha, B, Adams, M.W.W, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2005-05-26 | | Release date: | 2005-06-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Hypothetical Protein Pfu-631545-001 From Pyrococcus furiosus
To be Published
|
|
8GJG
 
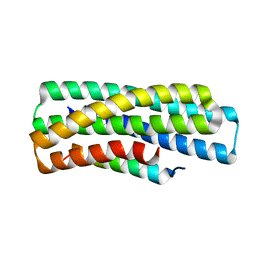 | | De novo design of high-affinity protein binders to bioactive helical peptides | | Descriptor: | gluc_A04_0005, gluc_A04_0005 Binder | | Authors: | Leung, P.J.Y, Bera, A.K, Torres, S.V, Baker, D, Kang, A. | | Deposit date: | 2023-03-15 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | De novo design of high-affinity binders of bioactive helical peptides.
Nature, 626, 2024
|
|
6N4N
 
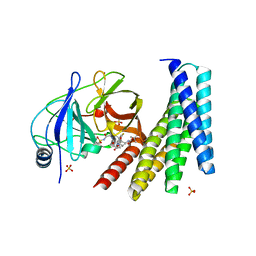 | | Crystal structure of the designed protein DNCR2/danoprevir/NS3a complex | | Descriptor: | (2R,6S,12Z,13aS,14aR,16aS)-6-[(tert-butoxycarbonyl)amino]-14a-[(cyclopropylsulfonyl)carbamoyl]-5,16-dioxo-1,2,3,5,6,7,8 ,9,10,11,13a,14,14a,15,16,16a-hexadecahydrocyclopropa[e]pyrrolo[1,2-a][1,4]diazacyclopentadecin-2-yl 4-fluoro-2H-isoindole-2-carboxylate, NS3 protease, Rosetta-designed danoprevir/NS3a complex reader 2, ... | | Authors: | Wang, Z, Foight, G.W, Baker, D, Maly, D.J. | | Deposit date: | 2018-11-19 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Multi-input chemical control of protein dimerization for programming graded cellular responses.
Nat.Biotechnol., 37, 2019
|
|
4ABJ
 
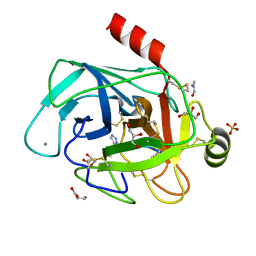 | | Co-complex structure of bovine trypsin with a modified Bowman-Birk inhibitor (IcA)SFTI-1(1,14), that was 1,5-disubstituted with 1,2,3- trizol to mimic a cis amide bond | | Descriptor: | CALCIUM ION, CATIONIC TRYPSIN, DIMETHYLFORMAMIDE, ... | | Authors: | Schmelz, S, Empting, M, Tischler, M, Nasu, D, Heinz, D, Kolmar, H. | | Deposit date: | 2011-12-08 | | Release date: | 2012-03-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Braces for the Peptide Backbone: Insights Into Structure-Activity Relation-Ships of Protease Inhibitor Mimics with Locked Amide Conformations
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
8DNT
 
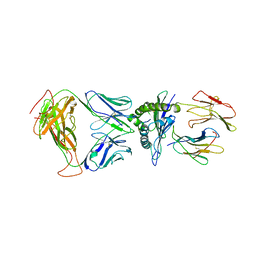 | | SARS-CoV-2 specific T cell receptor | | Descriptor: | Beta-2-microglobulin, MHC class I antigen alpha chain, Nucleoprotein, ... | | Authors: | Gallagher, D.T, Wu, D, Gowthaman, R, Pierce, B.G, Mariuzza, R.A, Weng, N.P. | | Deposit date: | 2022-07-11 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | SARS-CoV-2 infection establishes a stable and age-independent CD8 + T cell response against a dominant nucleocapsid epitope using restricted T cell receptors.
Nat Commun, 14, 2023
|
|
2XHH
 
 | | Circular permutation provides an evolutionary link between two families of calcium-dependent carbohydrate binding modules | | Descriptor: | (2S)-2-hydroxybutanedioic acid, CALCIUM ION, CARBOHYDRATE BINDING MODULE | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D, Ratnaparkhe, S, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-06-16 | | Release date: | 2010-07-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules
J.Biol.Chem., 285, 2010
|
|
8SY5
 
 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound the unnatural dS-BTP base pair in the active site | | Descriptor: | 2-oxo-2-hydroadenosine 5'-(tetrahydrogen triphosphate), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Shan, Z, Lyumkis, D, Oh, J, Wang, D. | | Deposit date: | 2023-05-24 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | A unified Watson-Crick geometry drives transcription of six-letter expanded DNA alphabets by E. coli RNA polymerase.
Nat Commun, 14, 2023
|
|
8SY6
 
 | | E. coli DNA-directed RNA polymerase transcription elongation complex bound the unnatural dB-UTP base pair in the active site | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Shan, Z, Lyumkis, D, Oh, J, Wang, D. | | Deposit date: | 2023-05-24 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | A unified Watson-Crick geometry drives transcription of six-letter expanded DNA alphabets by E. coli RNA polymerase.
Nat Commun, 14, 2023
|
|
2XRA
 
 | | crystal structure of the HK20 Fab in complex with a gp41 mimetic 5- Helix | | Descriptor: | HK20, HUMAN MONOCLONAL ANTIBODY HEAVY CHAIN, HUMAN MONOCLONAL ANTIBODY LIGHT CHAIN, ... | | Authors: | Sabin, C, Corti, D, Buzon, V, Seaman, M.S, Lutje Hulsik, D, Hinz, A, Vanzetta, F, Agatic, G, Silacci, C, Langedijk, J.P.M, Mainetti, L, Scarlatti, G, Sallusto, F, Weiss, R, Lanzavecchia, A, Weissenhorn, W. | | Deposit date: | 2010-09-13 | | Release date: | 2010-12-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and size-dependent neutralization properties of HK20, a human monoclonal antibody binding to the highly conserved heptad repeat 1 of gp41.
PLoS Pathog., 6, 2010
|
|
2XHJ
 
 | | Circular permutation provides an evolutionary link between two families of calcium-dependent carbohydrate binding modules. SeMet form of vCBM60. | | Descriptor: | CALCIUM ION, CALCIUM-DEPENDENT CARBOHYDRATE BINDING MODULE | | Authors: | Montanier, C, Flint, J.E, Bolam, D.N, Xie, H, Liu, Z, Rogowski, A, Weiner, D, Ratnaparkhe, S, Nurizzo, D, Roberts, S.M, Turkenburg, J.P, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2010-06-16 | | Release date: | 2010-07-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Circular Permutation Provides an Evolutionary Link between Two Families of Calcium-Dependent Carbohydrate Binding Modules
J.Biol.Chem., 285, 2010
|
|
