3X09
 
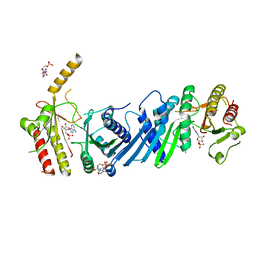 | | Crystal structure of PIP4KIIBETA F205L complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X04
 
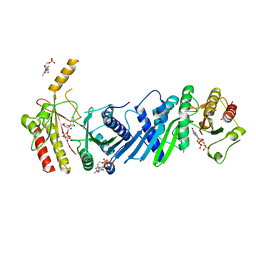 | | Crystal structure of PIP4KIIBETA complex with GMPPNP | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X02
 
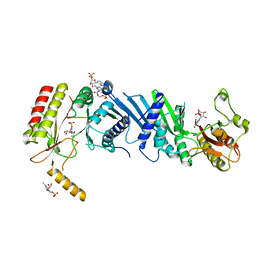 | | Crystal structure of PIP4KIIBETA complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
5EIG
 
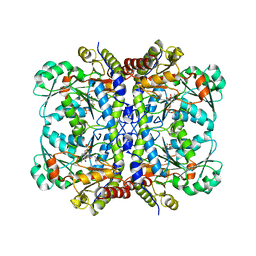 | |
3X0A
 
 | | Crystal structure of PIP4KIIBETA F205L complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
5Z50
 
 | | Crystal structure of PaCysB regulatory domain | | Descriptor: | Cys regulon transcriptional activator, GLYCEROL, SULFATE ION | | Authors: | Yang, C, Liang, H, Gan, J. | | Deposit date: | 2018-01-15 | | Release date: | 2019-01-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Molecular insights into the master regulator CysB-mediated bacterial virulence in Pseudomonas aeruginosa.
Mol.Microbiol., 111, 2019
|
|
8G9T
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | Descriptor: | AcrIC9, Cas11, Cas5, ... | | Authors: | Hu, C, Nam, K.H, Ke, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
8G9S
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | Descriptor: | AcrIC8, Cas11, Cas5, ... | | Authors: | Hu, C, Nam, K.H, Ke, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
8G9U
 
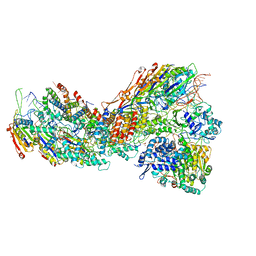 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | Descriptor: | CRISPR-associated protein, Csd1 family, Csd2 family, ... | | Authors: | Hu, C, Nam, K.H, Ke, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
5Z4Z
 
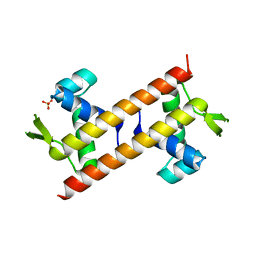 | |
6M2W
 
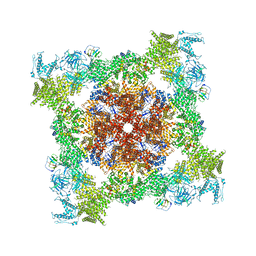 | | Structure of RyR1 (Ca2+/Caffeine/ATP/CaM1234/CHL) | | Descriptor: | 5-bromanyl-N-[4-chloranyl-2-methyl-6-(methylcarbamoyl)phenyl]-2-(3-chloranylpyridin-2-yl)pyrazole-3-carboxamide, ADENOSINE-5'-TRIPHOSPHATE, CAFFEINE, ... | | Authors: | Ma, R, Haji-Ghassemi, O, Ma, D, Lin, L, Samurkas, A, Van Petegem, F, Yuchi, Z. | | Deposit date: | 2020-03-01 | | Release date: | 2020-09-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for diamide modulation of ryanodine receptor.
Nat.Chem.Biol., 16, 2020
|
|
8GAF
 
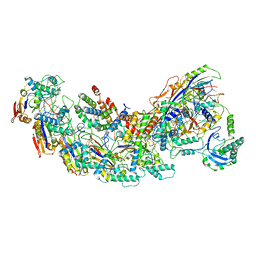 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | Descriptor: | Cas11, Cas5, Cas7, ... | | Authors: | Hu, C, Nam, K.H, Ke, A. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
8GAM
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | Descriptor: | Cas11, Cas5, Cas7, ... | | Authors: | Hu, C, Nam, K.H, Ke, A. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
8GAN
 
 | | Exploiting Activation and Inactivation Mechanisms in Type I-C CRISPR-Cas3 for Genome Editing Applications | | Descriptor: | Cas11, Cas5, Cas7, ... | | Authors: | Hu, C, Nam, K.H, Ke, A. | | Deposit date: | 2023-02-23 | | Release date: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Exploiting activation and inactivation mechanisms in type I-C CRISPR-Cas3 for genome-editing applications.
Mol.Cell, 84, 2024
|
|
8K4F
 
 | | DHODH in complex with compound A0 | | Descriptor: | 5-cyclopropyl-2-[1-[(2-fluorophenyl)methyl]pyrazolo[3,4-b]pyridin-3-yl]pyrimidin-4-amine, 6-[bis(oxidanyl)methyl]-5~{H}-pyrimidine-2,4-dione, ACETATE ION, ... | | Authors: | Jian, L, Sun, Q. | | Deposit date: | 2023-07-18 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Discovery and Optimization of Novel h DHODH Inhibitors for the Treatment of Inflammatory Bowel Disease.
J.Med.Chem., 66, 2023
|
|
7UD7
 
 | | Crystal structure of deoxygenated hemoglobin in complex with 5HMF-NO at 1.8 Angstrom | | Descriptor: | Hemoglobin subunit alpha, Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Donkor, A.K, Musayev, F.N, Safo, M.K. | | Deposit date: | 2022-03-18 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, and Antisickling Investigation of a Nitric Oxide-Releasing Prodrug of 5HMF for the Treatment of Sickle Cell Disease.
Biomolecules, 12, 2022
|
|
7UD8
 
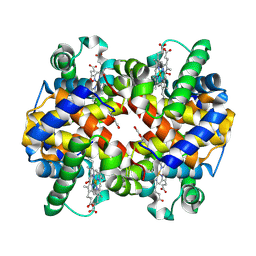 | | Crystal structure of carbon monoxy Hemoglobin in complex with 5HMF at 1.8 Angstrom | | Descriptor: | (5-methylfuran-2-yl)methanol, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Donkor, A.K, Musayev, F.N, Safo, M.S. | | Deposit date: | 2022-03-18 | | Release date: | 2022-03-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, Synthesis, and Antisickling Investigation of a Nitric Oxide-Releasing Prodrug of 5HMF for the Treatment of Sickle Cell Disease.
Biomolecules, 12, 2022
|
|
6M07
 
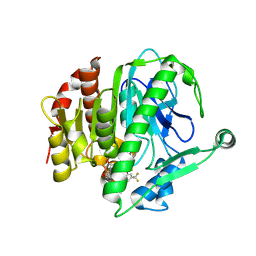 | | Crystal structure of Lp-PLA2 in complex with a novel covalent inhibitor | | Descriptor: | (2S)-2-[(E)-3-[2-(diethylamino)ethyl-[[4-[4-(trifluoromethyl)-2-[2,2,2-tris(fluoranyl)ethoxy]phenyl]phenyl]methyl]amino]-1-oxidanyl-3-oxidanylidene-prop-1-enyl]pyrrolidine-1-carboxylic acid, Platelet-activating factor acetylhydrolase | | Authors: | Hu, H.C, Xu, Y.C. | | Deposit date: | 2020-02-20 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Identification of Highly Selective Lipoprotein-Associated Phospholipase A2 (Lp-PLA2) Inhibitors by a Covalent Fragment-Based Approach.
J.Med.Chem., 63, 2020
|
|
4WY1
 
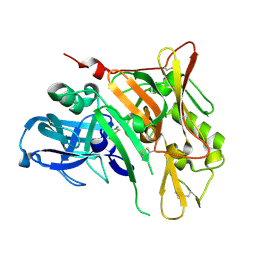 | | Crystal structure of human BACE-1 bound to Compound 24B | | Descriptor: | (4aR,8aS)-8a-(2,4-difluorophenyl)-4,4a,5,6,8,8a-hexahydropyrano[3,4-d][1,3]thiazin-2-amine, Beta-secretase 1 | | Authors: | Vajdos, F.F, Parris, K. | | Deposit date: | 2014-11-14 | | Release date: | 2015-03-04 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Discovery of a Series of Efficient, Centrally Efficacious BACE1 Inhibitors through Structure-Based Drug Design.
J.Med.Chem., 58, 2015
|
|
6M08
 
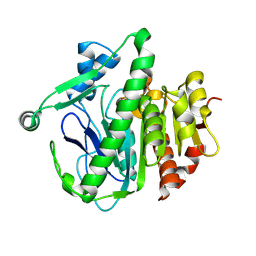 | | Crystal structure of Lp-PLA2 in complex with a novel covalent inhibitor. | | Descriptor: | (2S)-2-[(Z)-3-[2-(diethylamino)ethyl-[[4-[4-(trifluoromethyl)phenyl]phenyl]methyl]amino]-1-oxidanyl-3-oxidanylidene-prop-1-enyl]pyrrolidine-1-carboxylic acid, Platelet-activating factor acetylhydrolase | | Authors: | Hu, H.C, Xu, Y.C. | | Deposit date: | 2020-02-20 | | Release date: | 2020-12-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Identification of Highly Selective Lipoprotein-Associated Phospholipase A2 (Lp-PLA2) Inhibitors by a Covalent Fragment-Based Approach.
J.Med.Chem., 63, 2020
|
|
3VAX
 
 | | Crystal structure of DndA from streptomyces lividans | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, Putative uncharacterized protein dndA | | Authors: | Zhang, Z, Chen, F, Lin, K, Wu, G. | | Deposit date: | 2011-12-30 | | Release date: | 2013-01-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the cysteine desulfurase DndA from Streptomyces lividans which is involved in DNA phosphorothioation
Plos One, 7, 2012
|
|
4ZPV
 
 | | Structure of MERS-Coronavirus Spike Receptor-binding Domain (England1 Strain) in Complex with Vaccine-Elicited Murine Neutralizing Antibody D12 (Crystal Form 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, D12 Fab Heavy chain, D12 Fab light chain, ... | | Authors: | Joyce, M.G, Mascola, J.R, Graham, B.S, Kwong, P.D. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Evaluation of candidate vaccine approaches for MERS-CoV.
Nat Commun, 6, 2015
|
|
6JZO
 
 | | Structure of the mouse TRPC4 ion channel | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CHOLESTEROL HEMISUCCINATE, SODIUM ION, ... | | Authors: | Duan, J, Li, Z, Li, J, Zhang, J. | | Deposit date: | 2019-05-03 | | Release date: | 2020-10-21 | | Last modified: | 2021-05-05 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure of the mouse TRPC4 ion channel
To Be Published
|
|
3X0B
 
 | | Crystal structure of PIP4KIIBETA I368A complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
3X0C
 
 | | Crystal structure of PIP4KIIBETA I368A complex with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, Phosphatidylinositol 5-phosphate 4-kinase type-2 beta | | Authors: | Takeuchi, K, Lo, Y.H, Sumita, K, Senda, M, Terakawa, J, Dimitoris, A, Locasale, J.W, Sasaki, M, Yoshino, H, Zhang, Y, Kahoud, E.R, Takano, T, Yokota, T, Emerling, B, Asara, J.A, Ishida, T, Shimada, I, Daikoku, T, Cantley, L.C, Senda, T, Sasaki, A.T. | | Deposit date: | 2014-10-09 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Lipid Kinase PI5P4K beta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis
Mol.Cell, 61, 2016
|
|
