5FSM
 
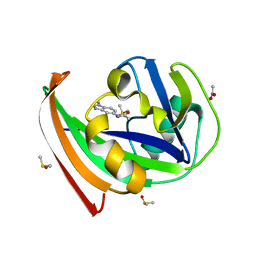 | | MTH1 substrate recognition: Complex with a methylbenzimidazolyl acetamide. | | Descriptor: | 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, ACETATE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Nissink, J.W.M, Bista, M, Breed, J, Carter, N, Embrey, K, Read, J, Phillips, C, Winter, J.J. | | Deposit date: | 2016-01-06 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Mth1 Substrate Recognition--an Example of Specific Promiscuity.
Plos One, 11, 2016
|
|
5FSO
 
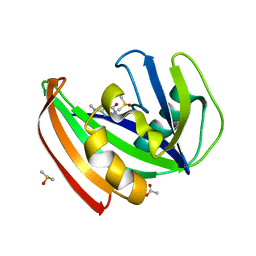 | | MTH1 substrate recognition: Complex with a methylaminopyrimidinedione. | | Descriptor: | 6-(METHYLAMINO)-1H-PYRIMIDINE-2,4-DIONE, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, ACETATE ION, ... | | Authors: | Nissink, J.W.M, Bista, M, Breed, J, Carter, N, Embrey, K, Read, J, Phillips, C, Winter, J.J. | | Deposit date: | 2016-01-06 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | MTH1 Substrate Recognition--An Example of Specific Promiscuity.
PLoS ONE, 11, 2016
|
|
5FSI
 
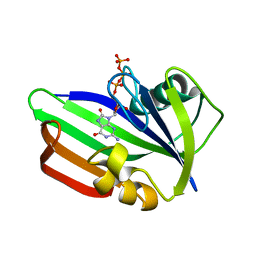 | | MTH1 substrate recognition: Complex with 8-oxo-dGTP. | | Descriptor: | 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE | | Authors: | Nissink, J.W.M, Bista, M, Breed, J, Carter, N, Embrey, K, Read, J, Phillips, C, Winter, J.J. | | Deposit date: | 2016-01-06 | | Release date: | 2016-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Mth1 Substrate Recognition--an Example of Specific Promiscuity.
Plos One, 11, 2016
|
|
5FSN
 
 | | MTH1 substrate recognition: Complex with a aminomethylpyrimidinyl oxypropanol. | | Descriptor: | 3-(2-AMINO-6-METHYL-PYRIMIDIN-4-YL)OXYPROPAN-1-OL, 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, ACETATE ION, ... | | Authors: | Nissink, J.W.M, Bista, M, Breed, J, Carter, N, Embrey, K, Read, J, Phillips, C, Winter, J.J. | | Deposit date: | 2016-01-06 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Mth1 Substrate Recognition--an Example of Specific Promiscuity.
Plos One, 11, 2016
|
|
5FSK
 
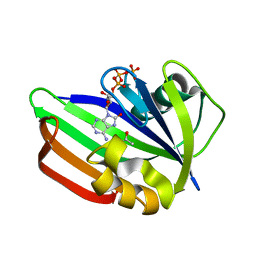 | | MTH1 substrate recognition: Complex with 8-oxo-dGTP. | | Descriptor: | 7,8-DIHYDRO-8-OXOGUANINE TRIPHOSPHATASE, 8-OXO-ADENOSINE-5'-TRIPHOSPHATE, ACETATE ION | | Authors: | Nissink, J.W.M, Bista, M, Breed, J, Carter, N, Embrey, K, Read, J, Phillips, C, Winter, J.J. | | Deposit date: | 2016-01-06 | | Release date: | 2016-07-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Mth1 Substrate Recognition--an Example of Specific Promiscuity.
Plos One, 11, 2016
|
|
5G55
 
 | | 3-Quinoline Carboxamides inhibitors of Pi3K | | Descriptor: | 6-cyano-4-[[(1R)-1-(4-methylphenyl)ethyl]amino]quinoline-3-carboxamide, PHOSPHATIDYLINOSITOL 4,5-BISPHOSPHATE 3-KINASE CATALYTIC SUBUNIT GAMMA ISOFORM, SULFATE ION | | Authors: | Edman, K, Phillips, C. | | Deposit date: | 2016-05-20 | | Release date: | 2016-08-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Discovery of Novel 3-Quinoline Carboxamides as Potent, Selective and Orally Bioavailable Inhibitors of Ataxia Telangiectasia Mutated (Atm) Kinase.
J.Med.Chem., 59, 2016
|
|
4CCL
 
 | | X-Ray structure of E. coli ycfD | | Descriptor: | 50S RIBOSOMAL PROTEIN L16 ARGININE HYDROXYLASE, MANGANESE (II) ION, SULFATE ION | | Authors: | McDonough, M.A, Ho, C.H, Kershaw, N.J, Schofield, C.J. | | Deposit date: | 2013-10-23 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4CSW
 
 | | Rhodothermus marinus YCFD-like ribosomal protein L16 Arginyl hydroxylase | | Descriptor: | CITRIC ACID, CUPIN 4 FAMILY PROTEIN, GLYCEROL, ... | | Authors: | McDonough, M.A, Sekirnik, R, Schofield, C.J. | | Deposit date: | 2014-03-11 | | Release date: | 2014-04-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.821 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4CUG
 
 | | Rhodothermus marinus YCFD-like ribosomal protein L16 Arginyl hydroxylase in complex substrate fragment | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 50S RIBOSOMAL PROTEIN L16, CUPIN 4 FAMILY PROTEIN, ... | | Authors: | McDonough, M.A, Sekirnik, R, Schofield, C.J. | | Deposit date: | 2014-03-18 | | Release date: | 2014-04-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 510, 2014
|
|
4M12
 
 | |
4M0Z
 
 | |
4M13
 
 | |
4DIQ
 
 | | Crystal Structure of human NO66 | | Descriptor: | Lysine-specific demethylase NO66, NICKEL (II) ION, PYRIDINE-2,4-DICARBOXYLIC ACID, ... | | Authors: | Vollmar, M, Krojer, T, Ng, S, Pilka, E, Bray, J, Pike, A.C.W, Filippakopoulos, P, Roos, A, Arrowsmith, C.H, Edwards, E, Bountra, C, von Delft, F, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-01-31 | | Release date: | 2012-03-07 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of human NO66
TO BE PUBLISHED
|
|
3TUB
 
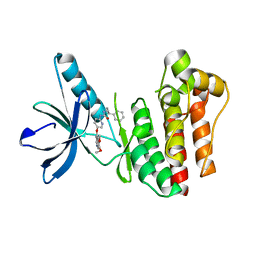 | |
2V95
 
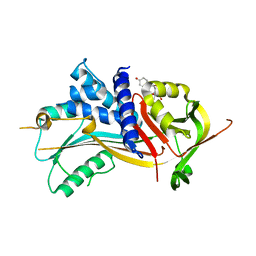 | | Structure of Corticosteroid-Binding Globulin in complex with Cortisol | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, CORTICOSTEROID-BINDING GLOBULIN | | Authors: | Klieber, M.A, Muller, Y.A. | | Deposit date: | 2007-08-21 | | Release date: | 2007-09-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Corticosteroid-Binding Globulin: Structural Basis for Steroid Transport and Proteinase-Triggered Release
J.Biol.Chem., 282, 2007
|
|
4LIT
 
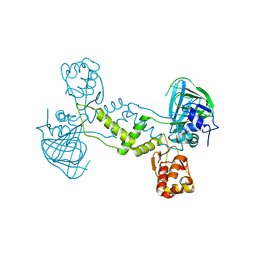 | |
4LIU
 
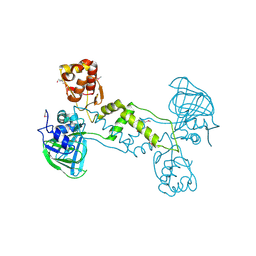 | | Structure of YcfD, a Ribosomal oxygenase from Escherichia coli. | | Descriptor: | 50S ribosomal protein L16 arginine hydroxylase, PHOSPHATE ION, TRIS(HYDROXYETHYL)AMINOMETHANE | | Authors: | Brissett, N.C, Doherty, A.J, Fox, G.C. | | Deposit date: | 2013-07-03 | | Release date: | 2014-05-14 | | Last modified: | 2014-07-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ribosomal oxygenases are structurally conserved from prokaryotes to humans.
Nature, 509, 2014
|
|
4LIV
 
 | |
3T5Z
 
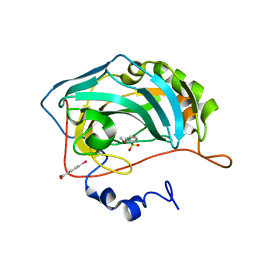 | | Crystal structure of the human carbonic anhydrase II in complex with N-methoxy-benzenesulfonamide | | Descriptor: | Carbonic anhydrase 2, MERCURIBENZOIC ACID, N-methoxybenzenesulfonamide, ... | | Authors: | Di Fiore, A, Maresca, A, Alterio, V, Supuran, C.T, De Simone, G. | | Deposit date: | 2011-07-28 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Carbonic anhydrase inhibitors: X-ray crystallographic studies for the binding of N-substituted benzenesulfonamides to human isoform II.
Chem.Commun.(Camb.), 47, 2011
|
|
3T5U
 
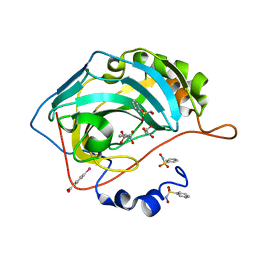 | | Crystal structure of the human carbonic anhydrase II in complex with N-hydroxy benzenesulfonamide | | Descriptor: | Carbonic anhydrase 2, GLYCEROL, MERCURIBENZOIC ACID, ... | | Authors: | Di Fiore, A, Maresca, A, Alterio, V, Supuran, C.T, De Simone, G. | | Deposit date: | 2011-07-28 | | Release date: | 2012-06-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Carbonic anhydrase inhibitors: X-ray crystallographic studies for the binding of N-substituted benzenesulfonamides to human isoform II.
Chem.Commun.(Camb.), 47, 2011
|
|
