7JM5
 
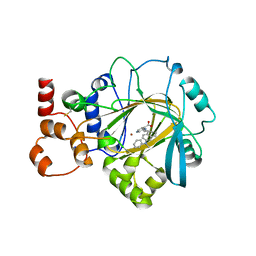 | | Crystal structure of KDM4B in complex with QC6352 | | Descriptor: | 3-[({(1R)-6-[methyl(phenyl)amino]-1,2,3,4-tetrahydronaphthalen-1-yl}methyl)amino]pyridine-4-carboxylic acid, Lysine-specific demethylase 4B, NICKEL (II) ION, ... | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-07-31 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Targeting KDM4 for treating PAX3-FOXO1-driven alveolar rhabdomyosarcoma.
Sci Transl Med, 14, 2022
|
|
8CMY
 
 | |
8HQS
 
 | | Cryo-EM structure of 8-subunit Smc5/6 head region | | Descriptor: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 3, Non-structural maintenance of chromosome element 4, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Tong, C, Wang, Z, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2022-12-14 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8I4U
 
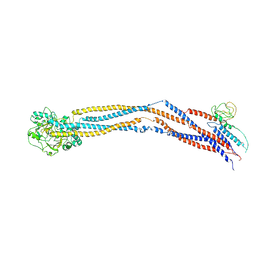 | | Cryo-EM structure of 5-subunit Smc5/6 hinge region | | Descriptor: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | Authors: | Qian, L, Jun, Z, Xiang, Z, Wang, Z, Tong, C, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.73 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8Y4U
 
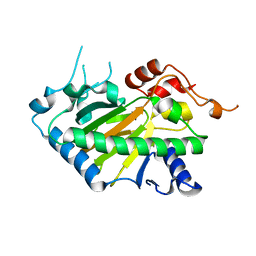 | | Crystal structure of a His1 from oryza sativa | | Descriptor: | FE (III) ION, Fe(II)/2-oxoglutarate-dependent oxygenase | | Authors: | Wang, N, Ma, J.M, Shibing, H, Beibei, Y, He, Z, Dandan, L. | | Deposit date: | 2024-01-30 | | Release date: | 2024-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of HPPD inhibitor sensitive protein from Oryza sativa.
Biochem.Biophys.Res.Commun., 704, 2024
|
|
2OBQ
 
 | | Discovery of the HCV NS3/4A Protease Inhibitor SCH503034. Key Steps in Structure-Based Optimization | | Descriptor: | Hepatitis C virus, ZINC ION | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-19 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC1
 
 | | Structure of the HCV NS3/4A Protease Inhibitor CVS4819 | | Descriptor: | (2S)-({N-[(3S)-3-({N-[(2S,4E)-2-ISOPROPYL-7-METHYLOCT-4-ENOYL]-L-LEUCYL}AMINO)-2-OXOHEXANOYL]GLYCYL}AMINO)(PHENYL)ACETI C ACID, Hepatitis C virus, ZINC ION | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OC0
 
 | | Structure of NS3 complexed with a ketoamide inhibitor SCh491762 | | Descriptor: | BETA-MERCAPTOETHANOL, Hepatitis C Virus, Hepatitis C virus, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-20 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
2OBO
 
 | | Structure of HEPATITIS C VIRAL NS3 protease domain complexed with NS4A peptide and ketoamide SCH476776 | | Descriptor: | BETA-MERCAPTOETHANOL, HCV NS3 protease, HCV NS4A peptide, ... | | Authors: | Prongay, A.J, Guo, Z, Yao, N, Fischmann, T, Strickland, C, Myers Jr, J, Weber, P.C, Malcolm, B, Beyer, B.M, Ingram, R, Pichardo, J, Hong, Z, Prosise, W.W, Ramanathan, L, Taremi, S.S, Yarosh-Tomaine, T, Zhang, R, Senior, M, Yang, R, Arasappan, A, Bennett, F, Bogen, S.F, Chen, K, Jao, E, Liu, Y, Love, R.G, Saksena, A.K, Venkatraman, S, Girijavallabhan, V, Njoroge, F.G, Madison, V. | | Deposit date: | 2006-12-19 | | Release date: | 2007-07-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of the HCV NS3/4A protease inhibitor (1R,5S)-N-[3-amino-1-(cyclobutylmethyl)-2,3-dioxopropyl]-3- [2(S)-[[[(1,1-dimethylethyl)amino]carbonyl]amino]-3,3-dimethyl-1-oxobutyl]- 6,6-dimethyl-3-azabicyclo[3.1.0]hexan-2(S)-carboxamide (Sch 503034) II. Key steps in structure-based optimization.
J.Med.Chem., 50, 2007
|
|
6WJH
 
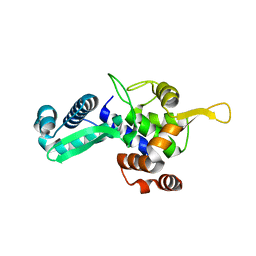 | |
6WWB
 
 | | Crystal Structure of the second bromodomain of human BRD2 in complex with the compound 3b | | Descriptor: | 1,2-ETHANEDIOL, 2-((S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-thieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-6-yl)-N-((1-(4-(2-((2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindolin-4-yl)amino)acetamido)butyl)-1H-1,2,3-triazol-4-yl)methyl)acetamide, Bromodomain-containing protein 2 | | Authors: | White, S.W, Yun, M. | | Deposit date: | 2020-05-08 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Phenyl-Glutarimides: Alternative Cereblon Binders for the Design of PROTACs.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
8I21
 
 | | Cryo-EM structure of 6-subunit Smc5/6 arm region | | Descriptor: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | Authors: | Jun, Z, Qian, L, Xiang, Z, Tong, C, Zhaoning, W, Duo, J, Zhenguo, C, Lanfeng, W. | | Deposit date: | 2023-01-13 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.02 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
8I4X
 
 | | Cryo-EM structure of 5-subunit Smc5/6 | | Descriptor: | E3 SUMO-protein ligase MMS21, Non-structural maintenance of chromosome element 5, Nse6, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Zhaoning, W, Tong, C, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 2024
|
|
1J4R
 
 | | FK506 BINDING PROTEIN COMPLEXED WITH FKB-001 | | Descriptor: | 1-[2,2-DIFLUORO-2-(3,4,5-TRIMETHOXY-PHENYL)-ACETYL]-PIPERIDINE-2-CARBOXYLIC ACID 4-PHENYL-1-(3-PYRIDIN-3-YL-PROPYL)-BUTYL ESTER, FK506-BINDING PROTEIN, GLYCEROL, ... | | Authors: | Sheriff, S. | | Deposit date: | 2001-10-29 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 2-Aryl-2,2-difluoroacetamide FKBP12 ligands: synthesis and X-ray structural studies.
Org.Lett., 3, 2001
|
|
8D1Y
 
 | |
8D24
 
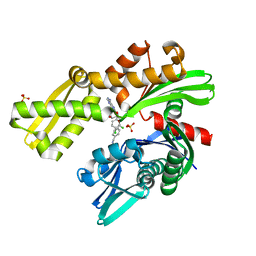 | | Crystal structure of Plasmodium falciparum GRP78-NBD in complex with VER155008 | | Descriptor: | 4-[[(2R,3S,4R,5R)-5-[6-amino-8-[(3,4-dichlorophenyl)methylamino]purin-9-yl]-3,4-dihydroxy-oxolan-2-yl]methoxymethyl]benzonitrile, Chaperone DnaK, SULFATE ION | | Authors: | Mrozek, A, Park, H.W. | | Deposit date: | 2022-05-27 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A non-traditional crystal-based compound screening method targeting the ATP binding site of Plasmodium falciparum GRP78 for identification of novel nucleoside analogues.
Front Mol Biosci, 9, 2022
|
|
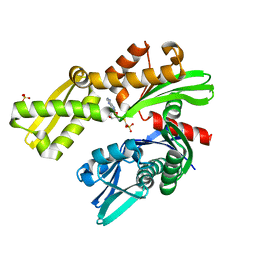
8D20
 
 | |
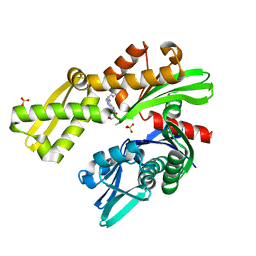
8D22
 
 | |
4YIF
 
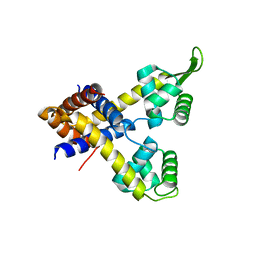 | | Crystal structure of Rv0880 | | Descriptor: | MarR family protein Rv0880 | | Authors: | Gao, Y.R, Feng, N, Li, D.F, Bi, L.J. | | Deposit date: | 2015-03-02 | | Release date: | 2015-06-10 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Structure of the MarR family protein Rv0880 from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
5D41
 
 | | EGFR kinase domain in complex with mutant selective allosteric inhibitor | | Descriptor: | (2R)-2-(1-oxo-1,3-dihydro-2H-isoindol-2-yl)-2-phenyl-N-(1,3-thiazol-2-yl)acetamide, Epidermal growth factor receptor, MAGNESIUM ION, ... | | Authors: | Yun, C.-H, Park, E, Eck, M.J. | | Deposit date: | 2015-08-07 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Overcoming EGFR(T790M) and EGFR(C797S) resistance with mutant-selective allosteric inhibitors.
Nature, 534, 2016
|
|
7EAM
 
 | | immune complex of SARS-CoV-2 RBD and cross-neutralizing antibody 7D6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike protein S1, ... | | Authors: | Li, T.T, Gu, Y, Li, S.W. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Cross-neutralizing antibodies bind a SARS-CoV-2 cryptic site and resist circulating variants.
Nat Commun, 12, 2021
|
|
7EAN
 
 | | immune complex of SARS-CoV-2 RBD and cross-neutralizing antibody 6D6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of SARS-CoV-2 cross-neutralizing mAb 6D6, Light chain of SARS-CoV-2 cross-neutralizing mAb 6D6, ... | | Authors: | Li, T.T, Gu, Y, Li, S.W. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Cross-neutralizing antibodies bind a SARS-CoV-2 cryptic site and resist circulating variants.
Nat Commun, 12, 2021
|
|
5Z3R
 
 | | Crystal Structure of Delta 5-3-Ketosteroid Isomerase from Mycobacterium sp. | | Descriptor: | Steroid delta-isomerase | | Authors: | Cheng, X.Y, Peng, F, Yang, F, Huang, Y.Q, Su, Z.D. | | Deposit date: | 2018-01-08 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structure-based reconstruction of a Mycobacterium hypothetical protein into an active Delta5-3-ketosteroid isomerase.
Biochim Biophys Acta Proteins Proteom, 1867, 2019
|
|
5V83
 
 | | Structure of DCN1 bound to NAcM-HIT | | Descriptor: | Lysozyme,DCN1-like protein 1 chimera, N-(1-benzylpiperidin-4-yl)-N'-[3-(trifluoromethyl)phenyl]urea | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
5V86
 
 | | Structure of DCN1 bound to NAcM-OPT | | Descriptor: | Lysozyme,DCN1-like protein 1, N-benzyl-N-(1-butylpiperidin-4-yl)-N'-(3,4-dichlorophenyl)urea | | Authors: | Guy, R.K, Schulman, B.A, Scott, D.C, Hammill, J.T. | | Deposit date: | 2017-03-21 | | Release date: | 2017-05-24 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.374 Å) | | Cite: | Blocking an N-terminal acetylation-dependent protein interaction inhibits an E3 ligase.
Nat. Chem. Biol., 13, 2017
|
|
