7MBN
 
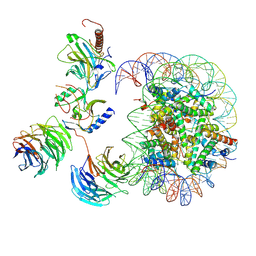 | | Cryo-EM structure of MLL1-NCP (H3K4M) complex, mode02 | | Descriptor: | DNA (146-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Dou, Y, Cho, U. | | Deposit date: | 2021-04-01 | | Release date: | 2021-12-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY | | Cite: | Regulation of MLL1 Methyltransferase Activity in Two Distinct Nucleosome Binding Modes.
Biochemistry, 61, 2022
|
|
7MBM
 
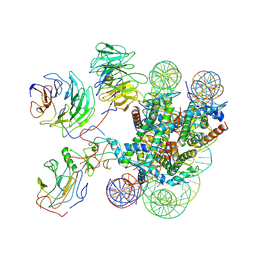 | | Cryo-EM structure of MLL1-NCP (H3K4M) complex, mode01 | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Dou, Y, Cho, U. | | Deposit date: | 2021-04-01 | | Release date: | 2021-12-29 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY | | Cite: | Regulation of MLL1 Methyltransferase Activity in Two Distinct Nucleosome Binding Modes.
Biochemistry, 61, 2022
|
|
4L5E
 
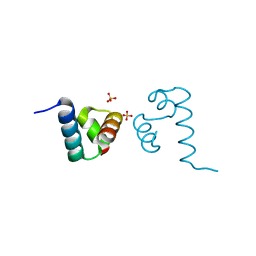 | | Crystal structure of A. aeolicus NtrC1 DNA binding domain | | Descriptor: | SULFATE ION, Transcriptional regulator (NtrC family) | | Authors: | Young, A, Maris, A.E, Vidangos, N.K, Hong, E, Pelton, J.G, Batchelor, J.D, Wemmer, D.E. | | Deposit date: | 2013-06-10 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structure, function, and tethering of DNA-binding domains in sigma (54) transcriptional activators.
Biopolymers, 99, 2013
|
|
1NFA
 
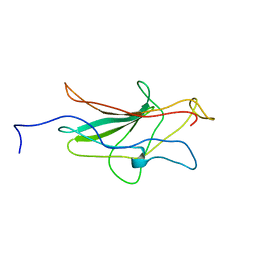 | | HUMAN TRANSCRIPTION FACTOR NFATC DNA BINDING DOMAIN, NMR, 10 STRUCTURES | | Descriptor: | HUMAN TRANSCRIPTION FACTOR NFATC1 | | Authors: | Wolfe, S.A, Zhou, P, Dotsch, V, Chen, L, You, A, Ho, S.N, Crabtree, G.R, Wagner, G, Verdine, G.L. | | Deposit date: | 1997-01-18 | | Release date: | 1997-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Unusual Rel-like architecture in the DNA-binding domain of the transcription factor NFATc.
Nature, 385, 1997
|
|
1SUX
 
 | | CRYSTALLOGRAPHIC ANALYSIS OF THE COMPLEX BETWEEN TRIOSEPHOSPHATE ISOMERASE FROM TRYPANOSOMA CRUZI AND 3-(2-benzothiazolylthio)-1-propanesulfonic acid | | Descriptor: | 3-(2-BENZOTHIAZOLYLTHIO)-1-PROPANESULFONIC ACID, SULFATE ION, Triosephosphate isomerase, ... | | Authors: | Tellez-Valencia, A, Olivares-Illana, V, Hernandez-Santoyo, A, Perez-Montfort, R, Costas, M, Rodriguez-Romero, A, Tuena De Gomez-Puyou, M, Gomez-Puyou, A. | | Deposit date: | 2004-03-26 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inactivation of triosephosphate isomerase from Trypanosoma cruzi by an agent that perturbs its dimer interface.
J.Mol.Biol., 341, 2004
|
|
1CI1
 
 | | CRYSTAL STRUCTURE OF TRIOSEPHOSPHATE ISOMERASE FROM TRYPANOSOMA CRUZI IN HEXANE | | Descriptor: | HEXANE, PROTEIN (TRIOSEPHOSPHATE ISOMERASE) | | Authors: | Gao, X.-G, Maldondo, E, Perez-Montfort, R, De Gomez-Puyou, M.T, Gomez-Puyou, A, Rodriguez-Romero, A. | | Deposit date: | 1999-04-06 | | Release date: | 1999-09-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of triosephosphate isomerase from Trypanosoma cruzi in hexane.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1TCD
 
 | | TRYPANOSOMA CRUZI TRIOSEPHOSPHATE ISOMERASE | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Maldonado, E, Soriano-Garcia, M, Cabrera, N, Garza-Ramos, G, Tuena De Gomez-Puyou, M, Gomez-Puyou, A, Perez-Montfort, R. | | Deposit date: | 1998-01-29 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Differences in the intersubunit contacts in triosephosphate isomerase from two closely related pathogenic trypanosomes.
J.Mol.Biol., 283, 1998
|
|
4HHP
 
 | | Crystal structure of triosephosphate isomerase from trypanosoma cruzi, mutant e105d | | Descriptor: | GLYCEROL, SULFATE ION, Triosephosphate isomerase, ... | | Authors: | Hernandez-Santoyo, A, Aguirre-Fuentes, Y, Torres-Larios, A, Gomez-Puyou, A, De Gomez-Puyou, M.T. | | Deposit date: | 2012-10-10 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Different contribution of conserved amino acids to the global properties of triosephosphate isomerases.
Proteins, 82, 2014
|
|
2VOM
 
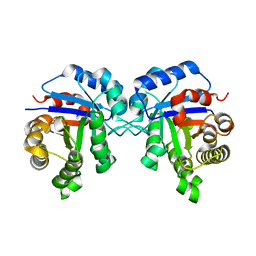 | | Structural basis of human triosephosphate isomerase deficiency. Mutation E104D and correlation to solvent perturbation. | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Rodriguez-Almazan, C, Arreola-Alemon, R, Rodriguez-Larrea, D, Aguirre-Lopez, B, de Gomez-Puyou, M.T, Perez-Montfort, R, Costas, M, Gomez-Puyou, A, Torres-Larios, A. | | Deposit date: | 2008-02-19 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Basis of Human Triosephosphate Isomerase Deficiency: Mutation E104D is Related to Alterations of a Conserved Water Network at the Dimer Interface.
J.Biol.Chem., 283, 2008
|
|
3TH6
 
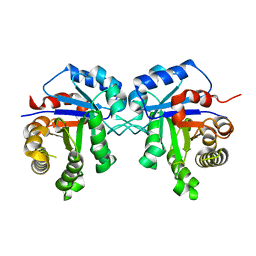 | | Crystal structure of Triosephosphate isomerase from Rhipicephalus (Boophilus) microplus. | | Descriptor: | Triosephosphate isomerase | | Authors: | Arreola, R, Rodriguez-Romero, A, Moraes, J, Gomez-Puyou, A, Perez-Montfort, R, Logullo, C. | | Deposit date: | 2011-08-18 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biochemical characterization of a recombinant triosephosphate isomerase from Rhipicephalus (Boophilus) microplus.
Insect Biochem.Mol.Biol., 41, 2011
|
|
4JEQ
 
 | | Different Contribution of Conserved Amino Acids to the Global Properties of Homologous Enzymes | | Descriptor: | DI(HYDROXYETHYL)ETHER, SULFATE ION, TRIOSEPHOSPHATE ISOMERASE, ... | | Authors: | Hernandez-Santoyo, A, Aguirre-Fuentes, Y, Torres-Larios, A, Gomez-Puyou, A, De Gomez-Puyou, M.T. | | Deposit date: | 2013-02-27 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Different contribution of conserved amino acids to the global properties of triosephosphate isomerases.
Proteins, 82, 2014
|
|
2JK2
 
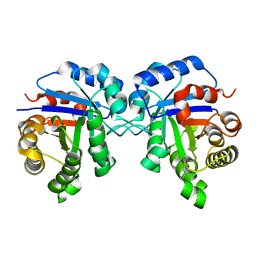 | | STRUCTURAL BASIS OF HUMAN TRIOSEPHOSPHATE ISOMERASE DEFICIENCY. CRYSTAL STRUCTURE OF THE WILD TYPE ENZYME. | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Rodriguez-Almazan, C, Arreola-Alemon, R, Rodriguez-Larrea, D, Aguirre-Lopez, B, De Gomez-Puyou, M.T, Perez-Montfort, R, Costas, M, Gomez-Puyou, A, Torres-Larios, A. | | Deposit date: | 2008-06-22 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis of Human Triosephosphate Isomerase Deficiency: Mutation E104D is Related to Alterations of a Conserved Water Network at the Dimer Interface.
J.Biol.Chem., 283, 2008
|
|
8FCI
 
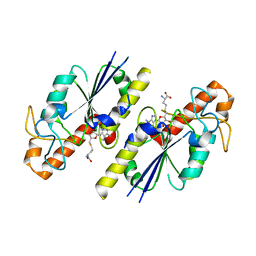 | | tRNA (N1G37) Methyltransferase (TrmD) of Mycobacterium avium complexed with S-Adenosyl homocysteine | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Lavallee, T, Mehta, K, Young, A, Cortes, J, Isaacson, B, Brylewski, J, Schlegel, F, Doti, L, Battaile, K, Stojanoff, V, Perez, A, Bolen, R. | | Deposit date: | 2022-12-01 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | tRNA (N1G37) Methyltransferase (TrmD) of Mycobacterium avium complexed with S-Adenosyl homocysteine
To Be Published
|
|
8FCH
 
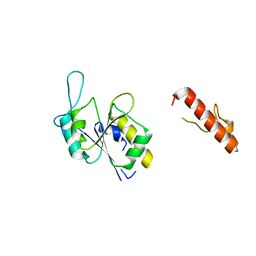 | | Apo Structure of (N1G37) Methyltransferase from Mycobacterium avium | | Descriptor: | tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Isaacson, B, Brylewski, J, Schlegel, F, Cortes, J, Lavallee, T, Mehta, K, Young, A, Doti, L, Battaile, K.P, Stojanoff, V, Perez, A, Bolen, R. | | Deposit date: | 2022-12-01 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.956 Å) | | Cite: | Apo structure of (N1G37) Methyltransferase from Mycobacterium avium.
To Be Published
|
|
6PWW
 
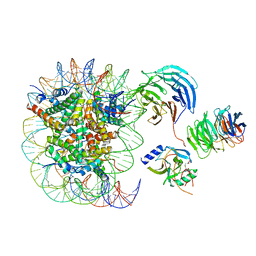 | | Cryo-EM structure of MLL1 in complex with RbBP5 and WDR5 bound to the nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
6PWX
 
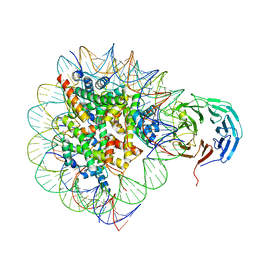 | | Cryo-EM structure of RbBP5 bound to the nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
6PWV
 
 | | Cryo-EM structure of MLL1 core complex bound to the nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
6W5N
 
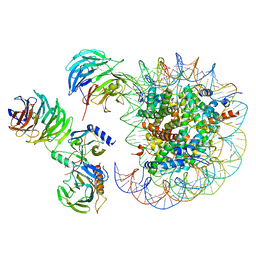 | | Cryo-EM structure of MLL1 in complex with RbBP5, WDR5, SET1, and ASH2L bound to the nucleosome (Class05) | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Lee, Y.T, Ayoub, A, Dou, Y, Cho, U. | | Deposit date: | 2020-03-13 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin.
Nat Commun, 12, 2021
|
|
6W5I
 
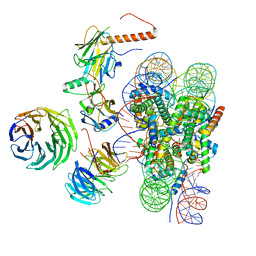 | | Cryo-EM structure of MLL1 in complex with RbBP5, WDR5, SET1, and ASH2L bound to the nucleosome (Class01) | | Descriptor: | DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Park, S.H, Lee, Y.T, Ayoub, A, Dou, Y, Cho, U. | | Deposit date: | 2020-03-13 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin.
Nat Commun, 12, 2021
|
|
6W5M
 
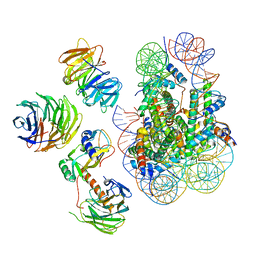 | | Cryo-EM structure of MLL1 in complex with RbBP5, WDR5, SET1, and ASH2L bound to the nucleosome (Class02) | | Descriptor: | DNA (147-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Lee, Y.T, Ayoub, A, Dou, Y, Cho, U. | | Deposit date: | 2020-03-13 | | Release date: | 2021-03-31 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin.
Nat Commun, 12, 2021
|
|
4L4U
 
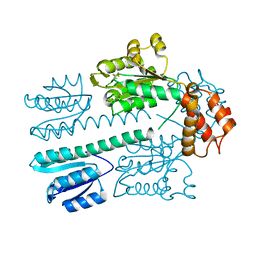 | | Crystal structure of construct containing A. aeolicus NtrC1 receiver, central and DNA binding domains | | Descriptor: | Transcriptional regulator (NtrC family) | | Authors: | Vidangos, N.K, Maris, A.E, Young, A, Hong, E, Pelton, J.G, Batchelor, J.D, Wemmer, D.E. | | Deposit date: | 2013-06-09 | | Release date: | 2013-08-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure, function, and tethering of DNA-binding domains in sigma (54) transcriptional activators.
Biopolymers, 99, 2013
|
|
1XWQ
 
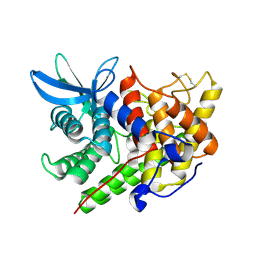 | | Structure Of A Cold-Adapted Family 8 Xylanase | | Descriptor: | beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, endo-1,4-beta-xylanase | | Authors: | De Vos, D, Collins, T, Hoyoux, A, Savvides, S.N, Gerday, C, Van Beeumen, J.J, Feller, G. | | Deposit date: | 2004-11-02 | | Release date: | 2005-10-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Study of the active site residues of a glycoside hydrolase family 8 xylanase
J.Mol.Biol., 354, 2005
|
|
1XWT
 
 | | Structure Of A Cold-Adapted Family 8 Xylanase | | Descriptor: | endo-1,4-beta-xylanase | | Authors: | De Vos, D, Collins, T, Hoyoux, A, Savvides, S.N, Gerday, C, Van Beeumen, J.J, Feller, G. | | Deposit date: | 2004-11-02 | | Release date: | 2005-10-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Study of the active site residues of a glycoside hydrolase family 8 xylanase
J.Mol.Biol., 354, 2005
|
|
1XW2
 
 | | Structure Of A Cold-Adapted Family 8 Xylanase | | Descriptor: | Endo-1,4-beta-Xylanase | | Authors: | Collins, T, De Vos, D, Hoyoux, A, Savvides, S.N, Gerday, C, Van Beeumen, J, Feller, G. | | Deposit date: | 2004-10-29 | | Release date: | 2005-10-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Study of the active site residues of a glycoside hydrolase family 8 xylanase
J.Mol.Biol., 354, 2005
|
|
2A8Z
 
 | | Structure Of A Cold-Adapted Family 8 Xylanase | | Descriptor: | endo-1,4-beta-xylanase | | Authors: | Collins, T, De Vos, D, Hoyoux, A, Savvides, S.N, Gerday, C, Van Beeumen, J, Feller, G. | | Deposit date: | 2005-07-10 | | Release date: | 2005-12-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Study of the active site residues of a glycoside hydrolase family 8 xylanase.
J.Mol.Biol., 354, 2005
|
|
