6BZ9
 
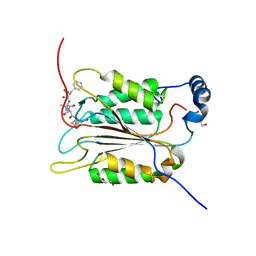 | | Crystal structure of human caspase-1 in complex with Ac-FLTD-CMK | | Descriptor: | Ac-FLTD-CMK, Caspase-1, DI(HYDROXYETHYL)ETHER | | Authors: | Xiao, T.S, Yang, J, Liu, Z, Wang, C, Yang, R. | | Deposit date: | 2017-12-22 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Mechanism of gasdermin D recognition by inflammatory caspases and their inhibition by a gasdermin D-derived peptide inhibitor.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5WPZ
 
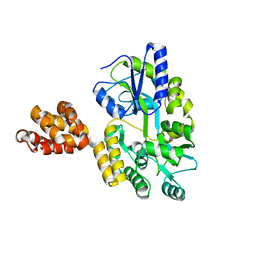 | | Crystal structure of MNDA PYD with MBP tag | | Descriptor: | MBP-hMNDA-PYD, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Jin, T.C, Xiao, T.S. | | Deposit date: | 2016-11-22 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of an expression system to enhance MBP-mediated crystallization
Sci Rep, 7, 2017
|
|
8T5P
 
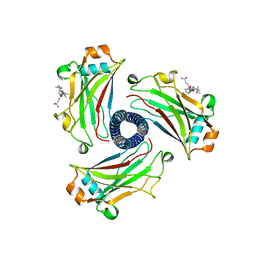 | |
8T5Q
 
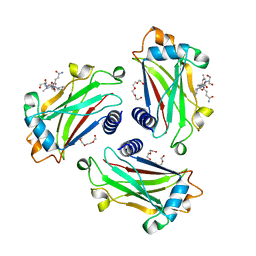 | |
6PZP
 
 | | Crystal structure of caspase-1 in complex with VX-765 | | Descriptor: | Caspase-1, N-(4-amino-3-chlorobenzene-1-carbonyl)-3-methyl-L-valyl-N-[(2S)-1-carboxy-3-oxopropan-2-yl]-L-prolinamide | | Authors: | Yang, J, Liu, Z, Xiao, T.S. | | Deposit date: | 2019-08-01 | | Release date: | 2020-08-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of caspase-1 in complex with VX-765
To Be Published
|
|
6UP4
 
 | |
6UP3
 
 | |
6UP2
 
 | |
6VIE
 
 | | Structure of caspase-1 in complex with gasdermin D | | Descriptor: | Caspase-1 subunit p10, Caspase-1 subunit p20, Gasdermin-D | | Authors: | Liu, Z, Xiao, T.S. | | Deposit date: | 2020-01-13 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Caspase-1 Engages Full-Length Gasdermin D through Two Distinct Interfaces That Mediate Caspase Recruitment and Substrate Cleavage.
Immunity, 53, 2020
|
|
6AO3
 
 | |
5WQ6
 
 | | Crystal Structure of hMNDA-PYD with MBP tag | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, MBP tagged hMNDA-PYD, ... | | Authors: | Jin, T.C, Xiao, T.S. | | Deposit date: | 2016-11-23 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Design of an expression system to enhance MBP-mediated crystallization
Sci Rep, 7, 2017
|
|
6N9N
 
 | | Crystal structure of murine GSDMD | | Descriptor: | Gasdermin-D | | Authors: | Liu, Z, Wang, C, Yang, J, Xiao, T.S. | | Deposit date: | 2018-12-03 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structures of the Full-Length Murine and Human Gasdermin D Reveal Mechanisms of Autoinhibition, Lipid Binding, and Oligomerization.
Immunity, 51, 2019
|
|
6N9O
 
 | | Crystal structure of human GSDMD | | Descriptor: | Gasdermin-D | | Authors: | Liu, Z, Wang, C, Yang, J, Xiao, T.S. | | Deposit date: | 2018-12-03 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structures of the Full-Length Murine and Human Gasdermin D Reveal Mechanisms of Autoinhibition, Lipid Binding, and Oligomerization.
Immunity, 51, 2019
|
|
4EO7
 
 | | Crystal structure of the TIR domain of human myeloid differentiation primary response protein 88. | | Descriptor: | MAGNESIUM ION, Myeloid differentiation primary response protein MyD88 | | Authors: | Snyder, G.A, Cirl, C, Jiang, J.S, Chen, P, Smith, T, Xiao, T.S. | | Deposit date: | 2012-04-13 | | Release date: | 2013-04-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Molecular mechanisms for the subversion of MyD88 signaling by TcpC from virulent uropathogenic Escherichia coli.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6NS7
 
 | |
6NRY
 
 | |
3VD8
 
 | | Crystal structure of human AIM2 PYD domain with MBP fusion | | Descriptor: | 1,2-ETHANEDIOL, Maltose-binding periplasmic protein, Interferon-inducible protein AIM2, ... | | Authors: | Jin, T.C, Perry, A, Smith, P, Xiao, T.S. | | Deposit date: | 2012-01-04 | | Release date: | 2013-01-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.0685 Å) | | Cite: | Structure of the Absent in Melanoma 2 (AIM2) Pyrin Domain Provides Insights into the Mechanisms of AIM2 Autoinhibition and Inflammasome Assembly.
J.Biol.Chem., 288, 2013
|
|
4QLC
 
 | | Crystal structure of chromatosome at 3.5 angstrom resolution | | Descriptor: | CITRIC ACID, DNA (167-mer), H5, ... | | Authors: | Jiang, J.S, Zhou, B.R, Xiao, T.S, Bai, Y.W. | | Deposit date: | 2014-06-11 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | Structural Mechanisms of Nucleosome Recognition by Linker Histones.
Mol.Cell, 33 Suppl 1, 2015
|
|
5H7Q
 
 | | Crystal structure of human MNDA PYD domain with MBP tag | | Descriptor: | ACETATE ION, MNDA PYD domain with MBP tag, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Jin, T.C, Xiao, T.S. | | Deposit date: | 2016-11-20 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Design of an expression system to enhance MBP-mediated crystallization
Sci Rep, 7, 2017
|
|
5H7N
 
 | | Crystal structure of human NLRP12-PYD with a MBP tag | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NLRP12-PYD with MBP tag, ... | | Authors: | Jin, T.C, Xiao, T.S. | | Deposit date: | 2016-11-19 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Design of an expression system to enhance MBP-mediated crystallization
Sci Rep, 7, 2017
|
|
4LQC
 
 | | The crystal structures of the Brucella protein TcpB and the TLR adaptor protein TIRAP show structural differences in microbial TIR mimicry. | | Descriptor: | TcpB | | Authors: | Snyder, G.A, Smith, P, Fresquez, T, Cirl, C, Jiang, J, Snyder, N, Luchetti, T, Miethke, T, Xiao, T.S. | | Deposit date: | 2013-07-17 | | Release date: | 2013-12-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the Toll/Interleukin-1 receptor (TIR) domains from the Brucella protein TcpB and host adaptor TIRAP reveal mechanisms of molecular mimicry.
J.Biol.Chem., 289, 2014
|
|
4LQD
 
 | | The crystal structures of the Brucella protein TcpB and the TLR adaptor protein TIRAP show structural differences in microbial TIR mimicry | | Descriptor: | GLYCEROL, Toll/interleukin-1 receptor domain-containing adapter protein | | Authors: | Snyder, G.A, Smith, P, Jiang, J, Xiao, T.S. | | Deposit date: | 2013-07-17 | | Release date: | 2013-12-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.451 Å) | | Cite: | Crystal structures of the Toll/Interleukin-1 receptor (TIR) domains from the Brucella protein TcpB and host adaptor TIRAP reveal mechanisms of molecular mimicry.
J.Biol.Chem., 289, 2014
|
|
4M6B
 
 | | Crystal structure of yeast Swr1-Z domain in complex with H2A.Z-H2B dimer | | Descriptor: | Chimera protein of Histone H2B.1 and Histone H2A.Z, Helicase SWR1 | | Authors: | Hong, J.J, Feng, H.Q, Wang, F, Ranjan, A, Chen, J.H, Jiang, J.S, Girlando, R, Xiao, T.S, Wu, C, Bai, Y.W. | | Deposit date: | 2013-08-09 | | Release date: | 2014-02-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Catalytic Subunit of the SWR1 Remodeler Is a Histone Chaperone for the H2A.Z-H2B Dimer.
Mol.Cell, 53, 2014
|
|
5Y3S
 
 | |
5Z7D
 
 | | p204HINab-dsDNA complex structure | | Descriptor: | DNA (5'-D(P*CP*CP*AP*TP*CP*AP*GP*AP*AP*AP*GP*AP*GP*AP*GP*C)-3'), Interferon-activable protein 204 | | Authors: | Jin, T, Jiang, J, Xiao, T.S. | | Deposit date: | 2018-01-28 | | Release date: | 2019-01-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Structural mechanism of DNA recognition by the p204 HIN domain.
Nucleic Acids Res., 2021
|
|
