2IOD
 
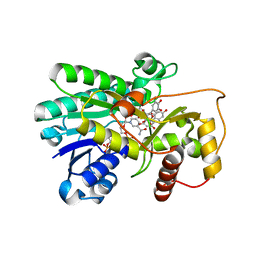 | | Binding of two substrate analogue molecules to dihydroflavonol-4-reductase alters the functional geometry of the catalytic site | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, Dihydroflavonol 4-reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petit, P, Langlois d'Estaintot, B, Granier, T, Gallois, B. | | Deposit date: | 2006-10-10 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Binding of two substrate analogue molecules to dihydroflavonol-4-reductase alters the functional geometry of the catalytic site
To be Published
|
|
4WJ0
 
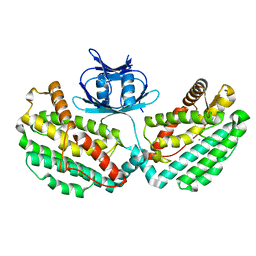 | | Structure of PH1245, a cas1 from Pyrococcus horikoshii | | Descriptor: | CHLORIDE ION, CRISPR-associated endonuclease Cas1 | | Authors: | Petit, P, Brown, G, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2014-09-29 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of PH1245, a cas1 from Pyrococcus horikoshii
To Be Published
|
|
4NO7
 
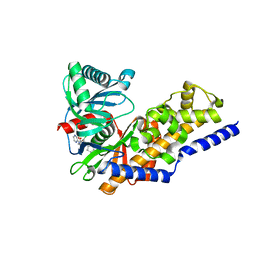 | | Human Glucokinase in complex with a nanomolar activator. | | Descriptor: | (2R)-2-[3-chloro-4-(methylsulfonyl)phenyl]-3-[(1R)-3-oxocyclopentyl]-N-(pyrazin-2-yl)propanamide, Glucokinase, alpha-D-glucopyranose | | Authors: | Petit, P, Ferry, G, Antoine, M, Boutin, J.A, Kotschy, A, Perron-Sierra, F, Mamelli, L, Vuillard, L. | | Deposit date: | 2013-11-19 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The active conformation of human glucokinase is not altered by allosteric activators.
Acta Crystallogr. D Biol. Crystallogr., 67, 2011
|
|
3R1W
 
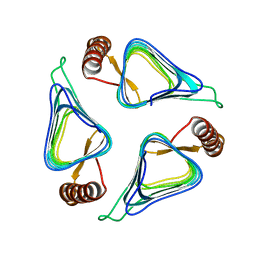 | | Crystal structure of a carbonic anhydrase from a crude oil degrading psychrophilic library | | Descriptor: | carbonic anhydrase | | Authors: | Petit, P, Xu, X, Cui, H, Brown, G, Dong, A, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2011-03-11 | | Release date: | 2011-03-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of a carbonic anhydrase from a crude oil degrading psychrophilic library
To be Published
|
|
3QYJ
 
 | |
3QYF
 
 | | Crystal structure of the CRISPR-associated protein SSO1393 from Sulfolobus solfataricus | | Descriptor: | CRISPR-ASSOCIATED PROTEIN, MAGNESIUM ION | | Authors: | Petit, P, Xu, X, Chang, C, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2011-03-03 | | Release date: | 2011-03-23 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the CRISPR-associated protein SSO1393 from Sulfolobus solfataricus
To be Published
|
|
2NNL
 
 | | Binding of two substrate analogue molecules to dihydroflavonol-4-reductase alters the functional geometry of the catalytic site | | Descriptor: | (2S)-2-(3,4-DIHYDROXYPHENYL)-5,7-DIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, Dihydroflavonol 4-reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petit, P, Langlois D'Estaintot, B, Granier, T, Gallois, B. | | Deposit date: | 2006-10-24 | | Release date: | 2007-11-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of two substrate analogue molecules to dihydroflavonol-4-reductase alters the functional geometry of the catalytic site
To be Published
|
|
2C29
 
 | | Structure of dihydroflavonol reductase from Vitis vinifera at 1.8 A. | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, DIHYDROFLAVONOL 4-REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petit, P, Granier, T, D'Estaintot, B.L, Hamdi, S, Gallois, B. | | Deposit date: | 2005-09-27 | | Release date: | 2006-10-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structure of Grape Dihydroflavonol 4-Reductase, a Key Enzyme in Flavonoid Biosynthesis.
J.Mol.Biol., 368, 2007
|
|
3FGU
 
 | | Catalytic complex of Human Glucokinase | | Descriptor: | Glucokinase, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Petit, P, Lagarde, A, Boutin, J.A, Ferry, G, Vuillard, L. | | Deposit date: | 2008-12-08 | | Release date: | 2009-12-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The active conformation of human glucokinase is not altered by allosteric activators
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3ID8
 
 | | Ternary complex of human pancreatic glucokinase crystallized with activator, glucose and AMP-PNP | | Descriptor: | 2-AMINO-4-FLUORO-5-[(1-METHYL-1H-IMIDAZOL-2-YL)SULFANYL]-N-(1,3-THIAZOL-2-YL)BENZAMIDE, Glucokinase, MAGNESIUM ION, ... | | Authors: | Petit, P, Gluais, L, Lagarde, A, Boutin, J.A, Ferry, G, Vuillard, L. | | Deposit date: | 2009-07-20 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The active conformation of human glucokinase is not altered by allosteric activators
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3F9M
 
 | | Human pancreatic glucokinase in complex with glucose and activator showing a mobile flap | | Descriptor: | 2-AMINO-4-FLUORO-5-[(1-METHYL-1H-IMIDAZOL-2-YL)SULFANYL]-N-(1,3-THIAZOL-2-YL)BENZAMIDE, Glucokinase, alpha-D-glucopyranose | | Authors: | Petit, P, Gluais, L, Lagarde, A, Vuillard, L, Boutin, J.A, Ferry, G. | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The active conformation of human glucokinase is not altered by allosteric activators
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3IDH
 
 | | Human pancreatic glucokinase in complex with glucose | | Descriptor: | Glucokinase, POTASSIUM ION, alpha-D-glucopyranose | | Authors: | Petit, P, Gluais, L, Lagarde, A, Boutin, J.A, Ferry, G, Vuillard, L. | | Deposit date: | 2009-07-21 | | Release date: | 2010-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | The active conformation of human glucokinase is not altered by allosteric activators
Acta Crystallogr.,Sect.D, 67, 2011
|
|
4XTK
 
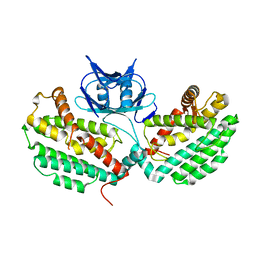 | | Structure of TM1797, a CAS1 protein from Thermotoga maritima | | Descriptor: | CRISPR-associated endonuclease Cas1 | | Authors: | Petit, P, Beloglazova, N, Skarina, T, Chang, C, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-01-23 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure and nuclease activity of tm1797, a cas1 protein from thermotoga maritima
To Be Published
|
|
4KUN
 
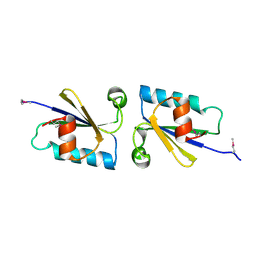 | | Crystal structure of Legionella pneumophila Lpp1115 / KaiB | | Descriptor: | Hypothetical protein Lpp1115 | | Authors: | Petit, P, Stogios, P.J, Stein, A, Wawrzak, Z, Skarina, T, Daniels, C, Di Leo, R, Buchrieser, C, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2013-05-22 | | Release date: | 2013-06-05 | | Last modified: | 2014-10-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Legionella pneumophila kai operon is implicated in stress response and confers fitness in competitive environments.
Environ Microbiol, 16, 2014
|
|
3DEL
 
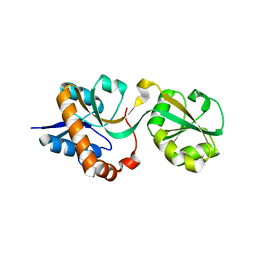 | |
3N26
 
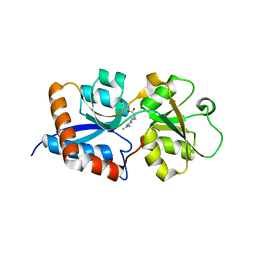 | | Cpn0482 : the arginine binding protein from the periplasm of chlamydia Pneumoniae | | Descriptor: | ARGININE, Amino acid ABC transporter, periplasmic amino acid-binding protein | | Authors: | Petit, P, Garcia, C, Vuillard, L, Soriani, M, Grandi, G, Marseilles Structural Genomics Program AFMB (MSGP), Marseilles Structural Genomics Program @ AFMB (MSGP) | | Deposit date: | 2010-05-17 | | Release date: | 2010-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exploiting antigenic diversity for vaccine design: the Chlamydia ArtJ paradigm.
J.Biol.Chem., 2010
|
|
3S8Y
 
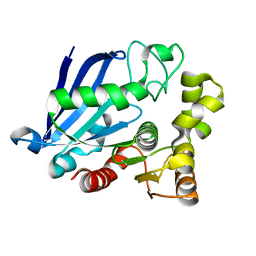 | | Bromide soaked structure of an esterase from the oil-degrading bacterium Oleispira antarctica | | Descriptor: | BROMIDE ION, Esterase APC40077 | | Authors: | Petit, P, Dong, A, Kagan, O, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2011-05-31 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and activity of the cold-active and anion-activated carboxyl esterase OLEI01171 from the oil-degrading marine bacterium Oleispira antarctica.
Biochem.J., 445, 2012
|
|
3PT1
 
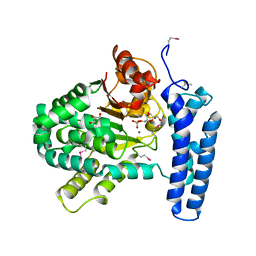 | | Structure of DUF89 from Saccharomyces cerevisiae co-crystallized with F6P. | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Petit, P, Xu, X, Cui, H, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2010-12-02 | | Release date: | 2010-12-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.773 Å) | | Cite: | Structure and activity of a DUF89 protein from Saccharomyces cerevisiae revealed a novel family of carbohydrate phosphatases
To be Published
|
|
3UMC
 
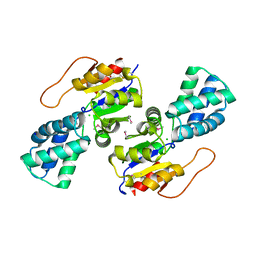 | | Crystal Structure of the L-2-Haloacid Dehalogenase PA0810 | | Descriptor: | CHLORIDE ION, SODIUM ION, haloacid dehalogenase | | Authors: | Petit, P, Chan, P.W.Y, Savchenko, A, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | Deposit date: | 2011-11-13 | | Release date: | 2012-11-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural adaptations of L-2-haloacid dehalogenases that enable hydrolytic defluorination
To be Published
|
|
3UMB
 
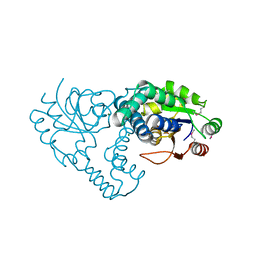 | | Crystal Structure of the L-2-Haloacid Dehalogenase RSc1362 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Petit, P, Chan, P.W.Y, Savchenko, A, Yakunin, A.F, Edwards, E.A, Pai, E.F. | | Deposit date: | 2011-11-12 | | Release date: | 2012-11-14 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural adaptations of L-2-haloacid dehalogenases that enable hydrolytic defluorination
To be Published
|
|
3S4L
 
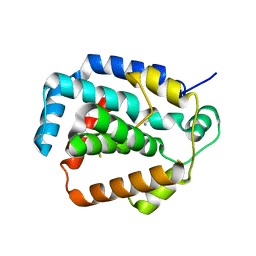 | | The CRISPR-associated Cas3 HD domain protein MJ0384 from Methanocaldococcus jannaschii | | Descriptor: | CALCIUM ION, CAS3 Metal dependent phosphohydrolase | | Authors: | Petit, P, Brown, G, Yakunin, A, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-05-19 | | Release date: | 2011-06-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and activity of the Cas3 HD nuclease MJ0384, an effector enzyme of the CRISPR interference.
Embo J., 30, 2011
|
|
3P48
 
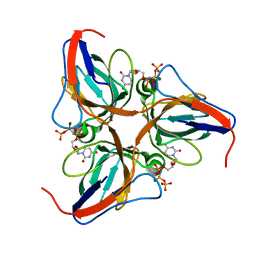 | | Structure of the yeast dUTPase DUT1 in complex with dUMPNPP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION | | Authors: | Petit, P, Singer, A.U, Evdokimova, E, Kudritska, M, Edwards, A.M, Yakunin, A.F, Savchenko, A, Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2010-10-06 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure and activity of the Saccharomyces cerevisiae dUTP pyrophosphatase DUT1, an essential housekeeping enzyme.
Biochem.J., 437, 2011
|
|
3C1T
 
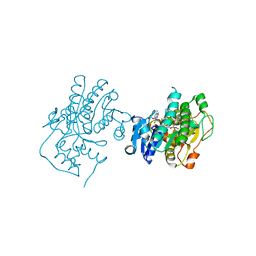 | | Binding of two substrate analogue molecules to dihydroflavonol 4-reductase alters the functional geometry of the catalytic site | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, dihydroflavonol 4-reductase | | Authors: | Trabelsi, N, Petit, P, Granier, T, Langlois d'Estaintot, B, Delrot, S, Gallois, B. | | Deposit date: | 2008-01-24 | | Release date: | 2008-02-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Structural evidence for the inhibition of grape dihydroflavonol 4-reductase by flavonols
Acta Crystallogr.,Sect.D, D64, 2008
|
|
3BXX
 
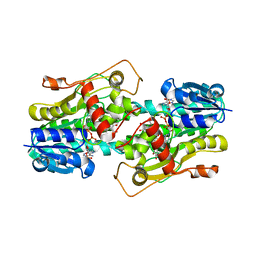 | | Binding of two substrate analogue molecules to dihydroflavonol 4-reductase alters the functional geometry of the catalytic site | | Descriptor: | 3,5,7,3',4'-PENTAHYDROXYFLAVONE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, dihydroflavonol 4-reductase | | Authors: | Trabelsi, N, Petit, P, Granier, T, Langlois d'Estaintot, B, Delrot, S, Gallois, B. | | Deposit date: | 2008-01-15 | | Release date: | 2008-10-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural evidence for the inhibition of grape dihydroflavonol 4-reductase by flavonols
Acta Crystallogr.,Sect.D, D64, 2008
|
|
3R0P
 
 | | Crystal structure of L-PSP putative endoribonuclease from uncultured organism | | Descriptor: | L-PSP putative endoribonuclease | | Authors: | Cuff, M.E, Petit, P, Xu, X, Cui, H, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2011-03-08 | | Release date: | 2011-03-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of L-PSP putative endoribonuclease from uncultured organism
To be Published
|
|
