1T09
 
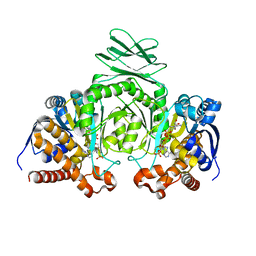 | | Crystal structure of human cytosolic NADP(+)-dependent isocitrate dehydrogenase in complex NADP | | Descriptor: | Isocitrate dehydrogenase [NADP] cytoplasmic, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Xu, X, Zhao, J, Peng, B, Huang, Q, Arnold, E, Ding, J. | | Deposit date: | 2004-04-08 | | Release date: | 2004-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity
J.Biol.Chem., 279, 2004
|
|
1T0L
 
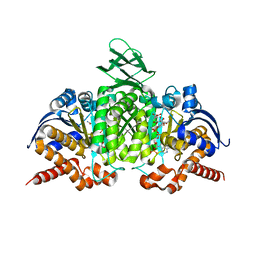 | | Crystal structure of human cytosolic NADP(+)-dependent isocitrate dehydrogenase in complex with NADP, isocitrate, and calcium(2+) | | Descriptor: | CALCIUM ION, ISOCITRIC ACID, Isocitrate dehydrogenase [NADP] cytoplasmic, ... | | Authors: | Xu, X, Zhao, J, Peng, B, Huang, Q, Arnold, E, Ding, J. | | Deposit date: | 2004-04-10 | | Release date: | 2004-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Structures of human cytosolic NADP-dependent isocitrate dehydrogenase reveal a novel self-regulatory mechanism of activity
J.Biol.Chem., 279, 2004
|
|
2HCR
 
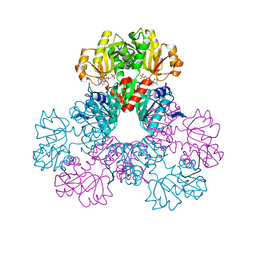 | | crystal structure of human phosphoribosyl pyrophosphate synthetase 1 in complex with AMP(ATP), cadmium and sulfate ion | | Descriptor: | ADENOSINE MONOPHOSPHATE, CADMIUM ION, Ribose-phosphate pyrophosphokinase I, ... | | Authors: | Li, S, Peng, B, Ding, J. | | Deposit date: | 2006-06-18 | | Release date: | 2006-10-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human phosphoribosylpyrophosphate synthetase 1 reveals a novel allosteric site
Biochem.J., 401, 2007
|
|
2H08
 
 | |
2H06
 
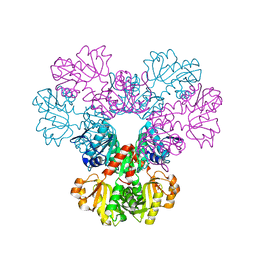 | |
2H07
 
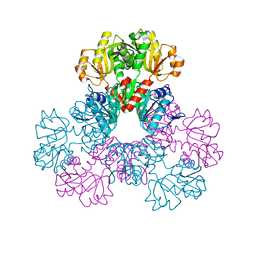 | |
7X8G
 
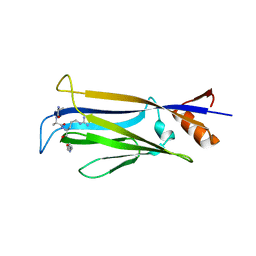 | |
7X8B
 
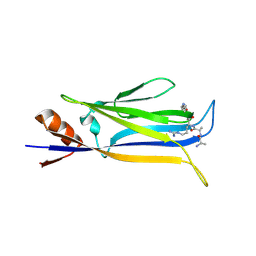 | |
7X8F
 
 | |
5LXP
 
 | | Human PARP14 (ARTD8), catalytic fragment in complex with inhibitor H5 | | Descriptor: | Poly [ADP-ribose] polymerase 14, ~{N}'-(3-aminocarbonylphenyl)-~{N}-[[1-[(2~{R})-2-phenylpropyl]-1,2,3-triazol-4-yl]methyl]pentanediamide | | Authors: | Karlberg, T, Thorsell, A.G, Schuler, H. | | Deposit date: | 2016-09-22 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Small Molecule Microarray Based Discovery of PARP14 Inhibitors.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5LYH
 
 | | Human PARP14 (ARTD8), catalytic fragment in complex with inhibitor H10 | | Descriptor: | 3-[2-[4-[2-[[4-[(3-aminocarbonylphenyl)amino]-4-oxidanylidene-butanoyl]amino]ethyl]-1,2,3-triazol-1-yl]ethylsulfamoyl]benzoic acid, Poly [ADP-ribose] polymerase 14 | | Authors: | Karlberg, T, Thorsell, A.G, Schuler, H. | | Deposit date: | 2016-09-28 | | Release date: | 2016-12-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Small Molecule Microarray Based Discovery of PARP14 Inhibitors.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
8B35
 
 | |
8B32
 
 | |
8B3C
 
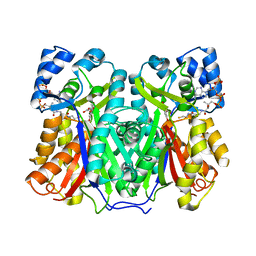 | | Chalcone synthase from Hordeum vulgare complexed with CoA and eriodictyol | | Descriptor: | (2S)-2-(3,4-DIHYDROXYPHENYL)-5,7-DIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, COENZYME A, Chalcone synthase 2 | | Authors: | Zhang, L, Groves, M.R. | | Deposit date: | 2022-09-16 | | Release date: | 2023-09-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering a Plant Polyketide Synthase for the Biosynthesis of Methylated Flavonoids.
J.Agric.Food Chem., 72, 2024
|
|
6VIO
 
 | |
3BKY
 
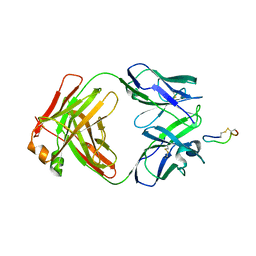 | | Crystal Structure of Chimeric Antibody C2H7 Fab in complex with a CD20 Peptide | | Descriptor: | B-lymphocyte antigen CD20, the Fab fragment of chimeric 2H7, heavy chain, ... | | Authors: | Du, J, Zhong, C, Ding, J. | | Deposit date: | 2007-12-07 | | Release date: | 2008-04-29 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of chimeric antibody C2H7 Fab in complex with a CD20 peptide
Mol.Immunol., 45, 2008
|
|
6FS1
 
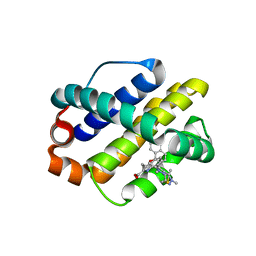 | | MCL1 in complex with an indole acid ligand | | Descriptor: | 1,2-ETHANEDIOL, 7-[3-[(1,5-dimethylpyrazol-3-yl)methylsulfanylmethyl]-1,5-dimethyl-pyrazol-4-yl]-3-(3-naphthalen-1-yloxypropyl)-1~{H}-indole-2-carboxylic acid, Induced myeloid leukemia cell differentiation protein Mcl-1 | | Authors: | Kasmirski, S, Hargreaves, D. | | Deposit date: | 2018-02-18 | | Release date: | 2018-12-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of Mcl-1-specific inhibitor AZD5991 and preclinical activity in multiple myeloma and acute myeloid leukemia.
Nat Commun, 9, 2018
|
|
5XP3
 
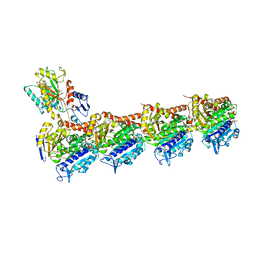 | | Crystal structure of apo T2R-TTL | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Yang, J, Wang, T, Chen, L. | | Deposit date: | 2017-05-31 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The compound millepachine and its derivatives inhibit tubulin polymerization by irreversibly binding to the colchicine-binding site in beta-tubulin.
J. Biol. Chem., 2018
|
|
6ZOQ
 
 | | Oestrogen receptor ligand binding domain in complex with compound 16 | | Descriptor: | Estrogen receptor, ~{N}-[4-[(6~{S},8~{R})-7-[(1-fluoranylcyclopropyl)methyl]-8-methyl-2,6,8,9-tetrahydropyrazolo[4,3-f]isoquinolin-6-yl]-3-methoxy-phenyl]-1-(3-fluoranylpropyl)azetidin-3-amine | | Authors: | Breed, J. | | Deposit date: | 2020-07-07 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of AZD9833, a Potent and Orally Bioavailable Selective Estrogen Receptor Degrader and Antagonist.
J.Med.Chem., 63, 2020
|
|
6ZOS
 
 | | Oestrogen receptor ligand binding domain in complex with compound 18 | | Descriptor: | 6-[(6~{S},8~{R})-7-[(1-fluoranylcyclopropyl)methyl]-8-methyl-2,6,8,9-tetrahydropyrazolo[4,3-f]isoquinolin-6-yl]-~{N}-[1-(3-fluoranylpropyl)azetidin-3-yl]pyridin-3-amine, Estrogen receptor | | Authors: | Breed, J. | | Deposit date: | 2020-07-07 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of AZD9833, a Potent and Orally Bioavailable Selective Estrogen Receptor Degrader and Antagonist.
J.Med.Chem., 63, 2020
|
|
6ZOR
 
 | | Oestrogen receptor ligand binding domain in complex with compound 28 | | Descriptor: | 6-[(6~{S},8~{R})-8-methyl-7-[2,2,2-tris(fluoranyl)ethyl]-3,6,8,9-tetrahydropyrazolo[4,3-f]isoquinolin-6-yl]-~{N}-(1-propylazetidin-3-yl)pyridin-3-amine, Estrogen receptor | | Authors: | Breed, J. | | Deposit date: | 2020-07-07 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery of AZD9833, a Potent and Orally Bioavailable Selective Estrogen Receptor Degrader and Antagonist.
J.Med.Chem., 63, 2020
|
|
3EOB
 
 | |
3EOA
 
 | |
3EO9
 
 | | Crystal structure the Fab fragment of Efalizumab | | Descriptor: | Efalizumab Fab fragment, heavy chain, light chain | | Authors: | Li, S, Ding, J. | | Deposit date: | 2008-09-26 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Efalizumab binding to the LFA-1 alphaL I domain blocks ICAM-1 binding via steric hindrance.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2OSL
 
 | | Crystal structure of Rituximab Fab in complex with an epitope peptide | | Descriptor: | B-lymphocyte antigen CD20, heavy chain of the Rituximab Fab fragment,heavy chain of the Rituximab Fab fragment, light chain of the Rituximab Fab fragment,light chain of the Rituximab Fab fragment | | Authors: | Du, J, Zhong, C, Ding, J. | | Deposit date: | 2007-02-06 | | Release date: | 2007-04-10 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for recognition of CD20 by therapeutic antibody Rituximab
J.Biol.Chem., 282, 2007
|
|
