3OE1
 
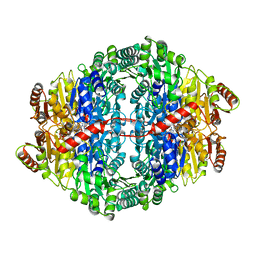 | | Pyruvate decarboxylase variant Glu473Asp from Z. mobilis in complex with reaction intermediate 2-lactyl-ThDP | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-CARBOXY-1-HYDROXYETHYL)-5-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Meyer, D, Neumann, P, Parthier, C, Tittmann, K. | | Deposit date: | 2010-08-12 | | Release date: | 2010-09-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.985 Å) | | Cite: | Double duty for a conserved glutamate in pyruvate decarboxylase: evidence of the participation in stereoelectronically controlled decarboxylation and in protonation of the nascent carbanion/enamine intermediate .
Biochemistry, 49, 2010
|
|
4FEG
 
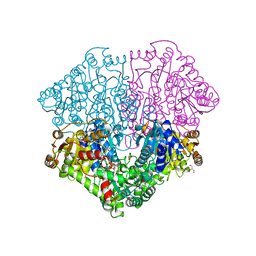 | | High-resolution structure of pyruvate oxidase in complex with reaction intermediate 2-hydroxyethyl-thiamin diphosphate carbanion-enamine, crystal A | | Descriptor: | 2-[(2E)-3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-HYDROXYETHYLIDENE)-4-METHYL-2,3-DIHYDRO-1,3-THIAZOL-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Meyer, D, Neumann, P, Koers, E, Sjuts, H, Luedtke, S, Sheldrick, G.M, Ficner, R, Tittmann, K. | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.09 Å) | | Cite: | Unexpected tautomeric equilibria of the carbanion-enamine intermediate in pyruvate oxidase highlight unrecognized chemical versatility of thiamin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4FEE
 
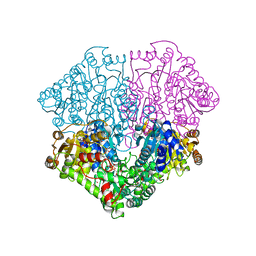 | | High-resolution structure of pyruvate oxidase in complex with reaction intermediate 2-hydroxyethyl-thiamin diphosphate carbanion-enamine, crystal B | | Descriptor: | 2-[(2E)-3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-HYDROXYETHYLIDENE)-4-METHYL-2,3-DIHYDRO-1,3-THIAZOL-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Meyer, D, Neumann, P, Koers, E, Sjuts, H, Luedtke, S, Sheldrick, G.M, Ficner, R, Tittmann, K. | | Deposit date: | 2012-05-30 | | Release date: | 2012-06-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Unexpected tautomeric equilibria of the carbanion-enamine intermediate in pyruvate oxidase highlight unrecognized chemical versatility of thiamin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1CDK
 
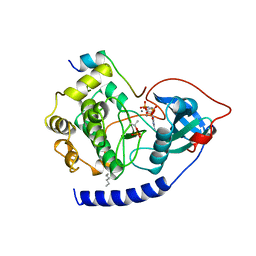 | | CAMP-DEPENDENT PROTEIN KINASE CATALYTIC SUBUNIT (E.C.2.7.1.37) (PROTEIN KINASE A) COMPLEXED WITH PROTEIN KINASE INHIBITOR PEPTIDE FRAGMENT 5-24 (PKI(5-24) ISOELECTRIC VARIANT CA) AND MN2+ ADENYLYL IMIDODIPHOSPHATE (MNAMP-PNP) AT PH 5.6 AND 7C AND 4C | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE, MANGANESE (II) ION, MYRISTIC ACID, ... | | Authors: | Bossemeyer, D, Engh, R.A, Kinzel, V, Ponstingl, H, Huber, R. | | Deposit date: | 1994-07-04 | | Release date: | 1995-10-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphotransferase and substrate binding mechanism of the cAMP-dependent protein kinase catalytic subunit from porcine heart as deduced from the 2.0 A structure of the complex with Mn2+ adenylyl imidodiphosphate and inhibitor peptide PKI(5-24).
EMBO J., 12, 1993
|
|
3KFK
 
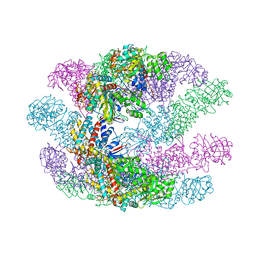 | | Crystal structures of a group II chaperonin from Methanococcus maripaludis | | Descriptor: | Chaperonin, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N, Meyer, D, Knee, K.M, Goulet, D.R, King, J.A, Frydman, J, Adams, P.D. | | Deposit date: | 2009-10-27 | | Release date: | 2010-06-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (6.003 Å) | | Cite: | Crystal structures of a group II chaperonin reveal the open and closed states associated with the protein folding cycle.
J.Biol.Chem., 285, 2010
|
|
3KFB
 
 | | Crystal structure of a group II chaperonin from Methanococcus maripaludis | | Descriptor: | Chaperonin, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N, Meyer, D, Knee, K.M, Goulet, D.R, King, J.A, Frydman, J, Adams, P.D. | | Deposit date: | 2009-10-27 | | Release date: | 2010-06-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of a group II chaperonin reveal the open and closed states associated with the protein folding cycle.
J.Biol.Chem., 285, 2010
|
|
3KFE
 
 | | Crystal structures of a group II chaperonin from Methanococcus maripaludis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperonin, MAGNESIUM ION, ... | | Authors: | Pereira, J.H, Ralston, C.Y, Douglas, N, Meyer, D, Knee, K.M, Goulet, D.R, King, J.A, Frydman, J, Adams, P.D. | | Deposit date: | 2009-10-27 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structures of a group II chaperonin reveal the open and closed states associated with the protein folding cycle.
J.Biol.Chem., 285, 2010
|
|
4JQ9
 
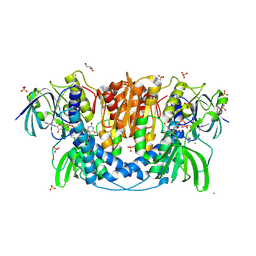 | | Dihydrolipoyl dehydrogenase of Escherichia coli pyruvate dehydrogenase complex | | Descriptor: | CHLORIDE ION, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tietzel, M, Neumann, P, Meyer, D, Ficner, R, Tittmann, K. | | Deposit date: | 2013-03-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Dihydrolipoyl dehydrogenase of Escherichia coli pyruvate dehydrogenase complex
TO BE PUBLISHED
|
|
2EZU
 
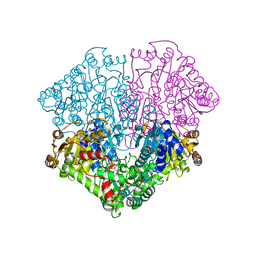 | | Pyruvate oxidase variant F479W in complex with reaction intermediate 2-acetyl-thiamin diphosphate | | Descriptor: | 2-ACETYL-THIAMINE DIPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Wille, G, Meyer, D, Steinmetz, A, Hinze, E, Golbik, R, Tittmann, K. | | Deposit date: | 2005-11-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The catalytic cycle of a thiamin diphosphate enzyme examined by cryocrystallography.
Nat.Chem.Biol., 2, 2006
|
|
2EZ9
 
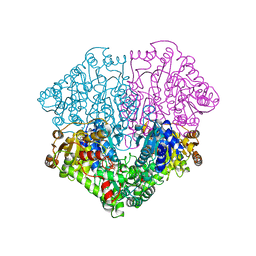 | | Pyruvate oxidase variant F479W in complex with reaction intermediate analogue 2-phosphonolactyl-thiamin diphosphate | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-{(1S)-1-HYDROXY-1-[(R)-HYDROXY(METHOXY)PHOSPHORYL]ETHYL}-5-(2-{[(S)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Wille, G, Meyer, D, Steinmetz, A, Hinze, E, Golbik, R, Tittmann, K. | | Deposit date: | 2005-11-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The catalytic cycle of a thiamin diphosphate enzyme examined by cryocrystallography.
Nat.Chem.Biol., 2, 2006
|
|
2EZT
 
 | | Pyruvate oxidase variant F479W in complex with reaction intermediate 2-hydroxyethyl-thiamin diphosphate | | Descriptor: | 2-[(2E)-3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-HYDROXYETHYLIDENE)-4-METHYL-2,3-DIHYDRO-1,3-THIAZOL-5-YL]ETHYL TRIHYDROGEN DIPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Wille, G, Meyer, D, Steinmetz, A, Hinze, E, Golbik, R, Tittmann, K. | | Deposit date: | 2005-11-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | The catalytic cycle of a thiamin diphosphate enzyme examined by cryocrystallography.
Nat.Chem.Biol., 2, 2006
|
|
2EZ8
 
 | | Pyruvate oxidase variant F479W in complex with reaction intermediate 2-lactyl-thiamin diphosphate | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-(1-CARBOXY-1-HYDROXYETHYL)-5-(2-{[HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Wille, G, Meyer, D, Steinmetz, A, Hinze, E, Golbik, R, Tittmann, K. | | Deposit date: | 2005-11-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.963 Å) | | Cite: | The catalytic cycle of a thiamin diphosphate enzyme examined by cryocrystallography.
Nat.Chem.Biol., 2, 2006
|
|
2EZ4
 
 | | Pyruvate oxidase variant F479W | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Wille, G, Meyer, D, Steinmetz, A, Hinze, E, Golbik, R, Tittmann, K. | | Deposit date: | 2005-11-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | The catalytic cycle of a thiamin diphosphate enzyme examined by cryocrystallography.
Nat.Chem.Biol., 2, 2006
|
|
3FZN
 
 | | Intermediate analogue in benzoylformate decarboxylase | | Descriptor: | 3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-2-{(S)-hydroxy[(R)-hydroxy(methoxy)phosphoryl]phenylmethyl}-5-(2-{[(R)-hydroxy(phosphonooxy)phosphoryl]oxy}ethyl)-4-methyl-1,3-thiazol-3-ium, Benzoylformate decarboxylase, CHLORIDE ION, ... | | Authors: | Bruning, M, Berheide, M, Meyer, D, Golbik, R, Bartunik, H, Liese, A, Tittmann, K. | | Deposit date: | 2009-01-26 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structural and kinetic studies on native intermediates and an intermediate analogue in benzoylformate decarboxylase reveal a least motion mechanism with an unprecedented short-lived predecarboxylation intermediate.
Biochemistry, 48, 2009
|
|
1STC
 
 | | CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH STAUROSPORINE | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE, PROTEIN KINASE INHIBITOR, STAUROSPORINE | | Authors: | Prade, L, Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1997-10-10 | | Release date: | 1998-02-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Staurosporine-induced conformational changes of cAMP-dependent protein kinase catalytic subunit explain inhibitory potential.
Structure, 5, 1997
|
|
1SZM
 
 | | DUAL BINDING MODE OF BISINDOLYLMALEIMIDE 2 TO PROTEIN KINASE A (PKA) | | Descriptor: | 3-(1H-INDOL-3-YL)-4-{1-[2-(1-METHYLPYRROLIDIN-2-YL)ETHYL]-1H-INDOL-3-YL}-1H-PYRROLE-2,5-DIONE, cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Gassel, M, Breitenlechner, C.B, Koenig, N, Huber, R, Engh, R.A, Bossemeyer, D. | | Deposit date: | 2004-04-06 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The protein kinase C inhibitor bisindolyl maleimide 2 binds with reversed orientations to different conformations of protein kinase a.
J.Biol.Chem., 279, 2004
|
|
2GNI
 
 | | PKA fivefold mutant model of Rho-kinase with inhibitor Fasudil (HA1077) | | Descriptor: | 5-(1,4-DIAZEPAN-1-SULFONYL)ISOQUINOLINE, cAMP-dependent protein kinase inhibitor alpha, cAMP-dependent protein kinase, ... | | Authors: | Bonn, S, Herrero, S, Breitenlechner, C.B, Engh, R.A, Gassel, M, Bossemeyer, D. | | Deposit date: | 2006-04-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural analysis of protein kinase A mutants with Rho-kinase inhibitor specificity
J.Biol.Chem., 281, 2006
|
|
2GNL
 
 | | PKA threefold mutant model of Rho-kinase with inhibitor H-1152P | | Descriptor: | (S)-2-METHYL-1-[(4-METHYL-5-ISOQUINOLINE)SULFONYL]-HOMOPIPERAZINE, cAMP-dependent protein kinase inhibitor alpha, cAMP-dependent protein kinase, ... | | Authors: | Bonn, S, Herrero, S, Breitenlechner, C.B, Engh, R.A, Gassel, M, Bossemeyer, D. | | Deposit date: | 2006-04-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis of protein kinase A mutants with Rho-kinase inhibitor specificity
J.Biol.Chem., 281, 2006
|
|
2GNF
 
 | | Protein kinase A fivefold mutant model of Rho-kinase with Y-27632 | | Descriptor: | (R)-TRANS-4-(1-AMINOETHYL)-N-(4-PYRIDYL) CYCLOHEXANECARBOXAMIDE, cAMP-dependent protein kinase inhibitor alpha, cAMP-dependent protein kinase, ... | | Authors: | Bonn, S, Herrero, S, Breitenlechner, C.B, Engh, R.A, Gassel, M, Bossemeyer, D. | | Deposit date: | 2006-04-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structural analysis of protein kinase A mutants with Rho-kinase inhibitor specificity
J.Biol.Chem., 281, 2006
|
|
2GNG
 
 | | Protein kinase A fivefold mutant model of Rho-kinase | | Descriptor: | cAMP-dependent protein kinase inhibitor alpha, cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Bonn, S, Herrero, S, Breitenlechner, C.B, Engh, R.A, Gassel, M, Bossemeyer, D. | | Deposit date: | 2006-04-10 | | Release date: | 2006-05-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural analysis of protein kinase A mutants with Rho-kinase inhibitor specificity
J.Biol.Chem., 281, 2006
|
|
3BWJ
 
 | | Complex of PKA with the bisubstrate protein kinase inhibitor lead compound Arc-1034 | | Descriptor: | (2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-N-(6-{[(1R)-4-carbamimidamido-1-{[(1R)-4-carbamimidamido-1-carbamoylbutyl]carbamoyl}butyl]amino}-6-oxohexyl)-3,4-dihydroxytetrahydrofuran-2-carboxamide, cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Lavogina, D, Koenig, N, Uri, A, Bossemeyer, D. | | Deposit date: | 2008-01-09 | | Release date: | 2009-02-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural analysis of ARC-type inhibitor (ARC-1034) binding to protein kinase A catalytic subunit and rational design of bisubstrate analogue inhibitors of basophilic protein kinases.
J.Med.Chem., 52, 2009
|
|
1YDR
 
 | | STRUCTURE OF CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH H7 PROTEIN KINASE INHIBITOR 1-(5-ISOQUINOLINESULFONYL)-2-METHYLPIPERAZINE | | Descriptor: | 1-(5-ISOQUINOLINESULFONYL)-2-METHYLPIPERAZINE, C-AMP-DEPENDENT PROTEIN KINASE, PROTEIN KINASE INHIBITOR PEPTIDE | | Authors: | Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1996-07-24 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity.
J.Biol.Chem., 271, 1996
|
|
1YDT
 
 | | STRUCTURE OF CAMP-DEPENDENT PROTEIN KINASE, ALPHA-CATALYTIC SUBUNIT IN COMPLEX WITH H89 PROTEIN KINASE INHIBITOR N-[2-(4-BROMOCINNAMYLAMINO)ETHYL]-5-ISOQUINOLINE | | Descriptor: | C-AMP-DEPENDENT PROTEIN KINASE, N-[2-(4-BROMOCINNAMYLAMINO)ETHYL]-5-ISOQUINOLINE SULFONAMIDE, PROTEIN KINASE INHIBITOR PEPTIDE | | Authors: | Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1996-07-24 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity.
J.Biol.Chem., 271, 1996
|
|
1YDS
 
 | | Structure of CAMP-dependent protein kinase, alpha-catalytic subunit in complex with H8 protein kinase inhibitor [N-(2-methylamino)ethyl]-5-isoquinolinesulfonamide | | Descriptor: | C-AMP-DEPENDENT PROTEIN KINASE, N-[2-(METHYLAMINO)ETHYL]-5-ISOQUINOLINESULFONAMIDE, PROTEIN KINASE INHIBITOR PEPTIDE | | Authors: | Engh, R.A, Girod, A, Kinzel, V, Huber, R, Bossemeyer, D. | | Deposit date: | 1996-07-24 | | Release date: | 1997-04-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of catalytic subunit of cAMP-dependent protein kinase in complex with isoquinolinesulfonyl protein kinase inhibitors H7, H8, and H89. Structural implications for selectivity.
J.Biol.Chem., 271, 1996
|
|
3AG9
 
 | | Complex of PKA with the bisubstrate protein kinase inhibitor ARC-1012 | | Descriptor: | (10R,20R,23R)-10-(4-aminobutyl)-1-[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]-20,23-bis(3-carbamimidamidopropyl)-1,8,11,18,21-pentaoxo-2,9,12,19,22-pentaazatetracosan-24-amide, cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Pflug, A, Lavogina, D, Uri, A, Engh, R.A, Bossemeyer, D. | | Deposit date: | 2010-03-26 | | Release date: | 2010-09-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diversity of bisubstrate binding modes of adenosine analogue-oligoarginine conjugates in protein kinase a and implications for protein substrate interactions.
J.Mol.Biol., 403, 2010
|
|
