6UQK
 
 | | Cryo-EM structure of type 3 IP3 receptor revealing presence of a self-binding peptide | | Descriptor: | ZINC ION, inositol 1,4,5-triphosphate receptor, type 3 | | Authors: | Azumaya, C.M, Linton, E.A, Risener, C.J, Nakagawa, T, Karakas, E. | | Deposit date: | 2019-10-20 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.77 Å) | | Cite: | Cryo-EM structure of human type-3 inositol triphosphate receptor reveals the presence of a self-binding peptide that acts as an antagonist.
J.Biol.Chem., 295, 2020
|
|
6CV9
 
 | |
2BSA
 
 | | Ferredoxin-Nadp Reductase (Mutation: Y 303 S) complexed with NADP | | Descriptor: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Maya, C.M, Hermoso, J.A, Perez-Dorado, I, Tejero, J, Julvez, M.M, Gomez-Moreno, C, Medina, M. | | Deposit date: | 2005-05-20 | | Release date: | 2005-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | C-Terminal Tyrosine of Ferredoxin-Nadp(+) Reductase in Hydride Transfer Processes with Nad(P)(+)/H.
Biochemistry, 44, 2005
|
|
4K1X
 
 | | Ferredoxin-NADP(H) Reductase mutant with Ala 266 replaced by Tyr (A266Y) and residues 267-272 deleted. | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADPH:ferredoxin reductase, SULFATE ION | | Authors: | Bortolotti, A, Sanchez-Azqueta, A, Maya, C.M, Velazquez-Campoy, A, Hermoso, J.A, Cortez, N. | | Deposit date: | 2013-04-06 | | Release date: | 2013-11-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The C-terminal extension of bacterial flavodoxin-reductases: Involvement in the hydride transfer mechanism from the coenzyme.
Biochim.Biophys.Acta, 1837, 2013
|
|
8CX0
 
 | | Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC monomeric complex | | Descriptor: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, Elongin-B, ... | | Authors: | Li, Y, Langley, C, Azumaya, C.M, Echeverria, I, Chesarino, N.M, Emerman, M, Cheng, Y, Gross, J.D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The structural basis for HIV-1 Vif antagonism of human APOBEC3G.
Nature, 615, 2023
|
|
8CX2
 
 | | Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC dimeric complex in State 2 | | Descriptor: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, Elongin-B, ... | | Authors: | Li, Y, Langley, C, Azumaya, C.M, Echeverria, I, Chesarino, N.M, Emerman, M, Cheng, Y, Gross, J.D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The structural basis for HIV-1 Vif antagonism of human APOBEC3G.
Nature, 615, 2023
|
|
8CX1
 
 | | Cryo-EM structure of human APOBEC3G/HIV-1 Vif/CBFbeta/ELOB/ELOC dimeric complex in State 1 | | Descriptor: | Core-binding factor subunit beta, DNA dC->dU-editing enzyme APOBEC-3G, Elongin-B, ... | | Authors: | Li, Y, Langley, C, Azumaya, C.M, Echeverria, I, Chesarino, N.M, Emerman, M, Cheng, Y, Gross, J.D. | | Deposit date: | 2022-05-19 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structural basis for HIV-1 Vif antagonism of human APOBEC3G.
Nature, 615, 2023
|
|
6W4S
 
 | | Structure of apo human ferroportin in lipid nanodisc | | Descriptor: | Fab45D8 Heavy Chain, Fab45D8 Light Chain, Solute carrier family 40 member 1 | | Authors: | Billesboelle, C.B, Azumaya, C.M, Gonen, S, Powers, A, Kretsch, R.C, Schneider, S, Arvedson, T, Dror, R.O, Cheng, Y, Manglik, A. | | Deposit date: | 2020-03-11 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of hepcidin-bound ferroportin reveals iron homeostatic mechanisms.
Nature, 586, 2020
|
|
6WBV
 
 | | Structure of human ferroportin bound to hepcidin and cobalt in lipid nanodisc | | Descriptor: | (1S)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PENTANOYLOXY)METHYL]ETHYL OCTANOATE, COBALT (II) ION, Fab45D8 Heavy Chain, ... | | Authors: | Billesboelle, C.B, Azumaya, C.M, Gonen, S, Powers, A, Kretsch, R.C, Schneider, S, Arvedson, T, Dror, R.O, Cheng, Y, Manglik, A. | | Deposit date: | 2020-03-27 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structure of hepcidin-bound ferroportin reveals iron homeostatic mechanisms.
Nature, 586, 2020
|
|
6W4V
 
 | | Structure of anti-ferroportin Fab45D8 | | Descriptor: | 1,2-ETHANEDIOL, Fab45D8 Heavy Chain, Fab45D8 Light Chain | | Authors: | Billesboelle, C.B, Azumaya, C.M, Gonen, S, Powers, A, Kretsch, R.C, Schneider, S, Arvedson, T, Dror, R.O, Cheng, Y, Manglik, A. | | Deposit date: | 2020-03-11 | | Release date: | 2020-09-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of hepcidin-bound ferroportin reveals iron homeostatic mechanisms.
Nature, 586, 2020
|
|
6D7L
 
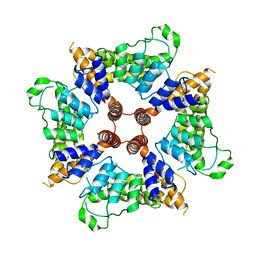 | |
6DJR
 
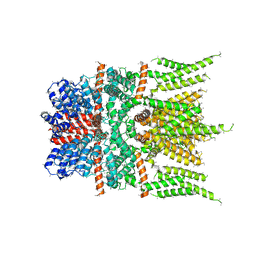 | |
6DJS
 
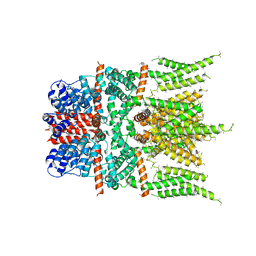 | |
7KKJ
 
 | | Structure of anti-SARS-CoV-2 Spike nanobody mNb6 | | Descriptor: | CHLORIDE ION, SULFATE ION, Synthetic nanobody mNb6 | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KKK
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody Nb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
7KKL
 
 | | SARS-CoV-2 Spike in complex with neutralizing nanobody mNb6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Schoof, M.S, Faust, B.F, Saunders, R.A, Sangwan, S, Rezelj, V, Hoppe, N, Boone, M, Billesboelle, C.B, Puchades, C, Azumaya, C.M, Kratochvil, H.T, Zimanyi, M, Desphande, I, Liang, J, Dickinson, S, Nguyen, H.C, Chio, C.M, Merz, G.E, Thompson, M.C, Diwanji, D, Schaefer, K, Anand, A.A, Dobzinski, N, Zha, B.S, Simoneau, C.R, Leon, K, White, K.M, Chio, U.S, Gupta, M, Jin, M, Li, F, Liu, Y, Zhang, K, Bulkley, D, Sun, M, Smith, A.M, Rizo, A.N, Moss, F, Brilot, A.F, Pourmal, S, Trenker, R, Pospiech, T, Gupta, S, Barsi-Rhyne, B, Belyy, V, Barile-Hill, A.W, Nock, S, Liu, Y, Krogan, N.J, Ralston, C.Y, Swaney, D.L, Garcia-Sastre, A, Ott, M, Vignuzzi, M, Walter, P, Manglik, A, QCRG Structural Biology Consortium | | Deposit date: | 2020-10-27 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike.
Science, 370, 2020
|
|
4R1Z
 
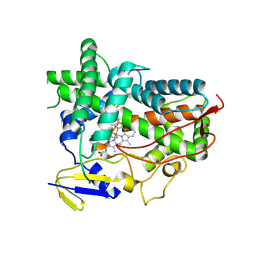 | | Zebra fish cytochrome P450 17A1 with Abiraterone | | Descriptor: | Abiraterone, Cyp17a1 protein, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2014-08-08 | | Release date: | 2014-12-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and Kinetic Basis of Steroid 17 alpha, 20-Lyase Activity in Teleost Fish Cytochrome P450 17A1 and Its Absence in Cytochrome P450 17A2.
J.Biol.Chem., 290, 2015
|
|
6UFI
 
 | | W96Y Oxalate Decarboxylase (Bacillus subtilis) | | Descriptor: | CHLORIDE ION, Cupin domain-containing protein, GLYCEROL, ... | | Authors: | Pastore, A.J, Burg, M.J, Twahir, U.T, Bruner, S.D, Angerhofer, A. | | Deposit date: | 2019-09-24 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Oxalate decarboxylase uses electron hole hopping for catalysis.
J.Biol.Chem., 297, 2021
|
|
8R38
 
 | | BIIG2 anti-integrin Fab | | Descriptor: | BIIG2 Fab, heavy chain, light chain, ... | | Authors: | Cordara, G, Heim, J.B, Johannesen, H, Krengel, U. | | Deposit date: | 2023-11-08 | | Release date: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural and functional characterization of integrin alpha5-targeting antibodies for anti-angiogenic therapy
To Be Published
|
|
8EM1
 
 | | Type IIS Restriction Endonuclease PaqCI, DNA Unbound | | Descriptor: | 1,2-ETHANEDIOL, PaqCI, DNA Unbound | | Authors: | Kennedy, M.A, Stoddard, B.L. | | Deposit date: | 2022-09-26 | | Release date: | 2023-03-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures, activity and mechanism of the Type IIS restriction endonuclease PaqCI.
Nucleic Acids Res., 51, 2023
|
|
8EPX
 
 | | Type IIS Restriction Endonuclease PaqCI, DNA bound | | Descriptor: | CALCIUM ION, DNA 1a, DNA 1b, ... | | Authors: | Kennedy, M.A, Stoddard, B.L. | | Deposit date: | 2022-10-06 | | Release date: | 2023-03-22 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structures, activity and mechanism of the Type IIS restriction endonuclease PaqCI.
Nucleic Acids Res., 51, 2023
|
|
4R20
 
 | | Zebra fish cytochrome P450 17A2 with Abiraterone | | Descriptor: | Abiraterone, Cytochrome P450 family 17 polypeptide 2, MERCURY (II) ION, ... | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2014-08-08 | | Release date: | 2014-12-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Structural and Kinetic Basis of Steroid 17 alpha, 20-Lyase Activity in Teleost Fish Cytochrome P450 17A1 and Its Absence in Cytochrome P450 17A2.
J.Biol.Chem., 290, 2015
|
|
4R21
 
 | | Zebra fish cytochrome P450 17A2 with Progesterone | | Descriptor: | Cytochrome P450 family 17 polypeptide 2, PROGESTERONE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Pallan, P.S, Egli, M. | | Deposit date: | 2014-08-08 | | Release date: | 2014-12-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Kinetic Basis of Steroid 17 alpha, 20-Lyase Activity in Teleost Fish Cytochrome P450 17A1 and Its Absence in Cytochrome P450 17A2.
J.Biol.Chem., 290, 2015
|
|
7KDT
 
 | |
7KOD
 
 | | Cryo-EM structure of heavy chain mouse apoferritin | | Descriptor: | Ferritin heavy chain | | Authors: | Sun, M, Azumaya, C, Tse, E, Frost, A, Southworth, D, Verba, K.A, Cheng, Y, Agard, D.A. | | Deposit date: | 2020-11-08 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (1.655 Å) | | Cite: | Practical considerations for using K3 cameras in CDS mode for high-resolution and high-throughput single particle cryo-EM.
J.Struct.Biol., 213, 2021
|
|
