8HGV
 
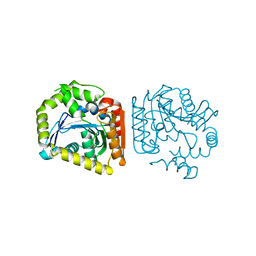 | |
7KE5
 
 | | Heavy chain ferritin with N-terminal EBNA1 epitope | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Epstein-Barr nuclear antigen 1,Ferritin heavy chain, FE (III) ION | | Authors: | Pederick, J.P, Bruning, J.B. | | Deposit date: | 2020-10-10 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Immunogenicity study of engineered ferritins with C- and N-terminus insertion of Epstein-Barr nuclear antigen 1 epitope.
Vaccine, 39, 2021
|
|
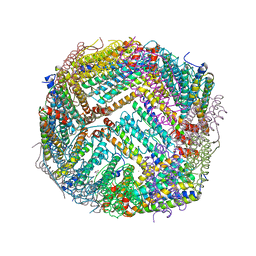
7KE3
 
 | | Heavy chain ferritin with C-terminal EBNA1 epitope | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE (III) ION, Ferritin heavy chain,Epstein-Barr nuclear antigen 1 | | Authors: | Pederick, J.L, Bruning, J.B. | | Deposit date: | 2020-10-10 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Immunogenicity study of engineered ferritins with C- and N-terminus insertion of Epstein-Barr nuclear antigen 1 epitope.
Vaccine, 39, 2021
|
|
8HLP
 
 | | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 (apo) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wei, Y, Yu, Z, Zhao, Y. | | Deposit date: | 2022-11-30 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural bases of inhibitory mechanism of Ca V 1.2 channel inhibitors.
Nat Commun, 15, 2024
|
|
2MP0
 
 | | Protein Phosphorylation upon a Fleeting Encounter | | Descriptor: | Glucose-specific phosphotransferase enzyme IIA component, PHOSPHITE ION, Phosphoenolpyruvate-protein phosphotransferase | | Authors: | Xing, Q, Yang, J, Huang, P, Zhang, W, Tang, C. | | Deposit date: | 2014-05-08 | | Release date: | 2014-08-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Visualizing an ultra-weak protein-protein interaction in phosphorylation signaling.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
8HMB
 
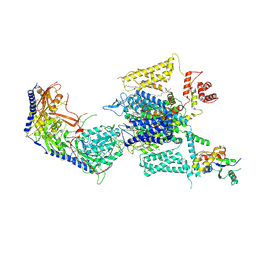 | | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 in complex with benidipine (BEN) | | Descriptor: | (3R)-1-benzylpiperidin-3-yl methyl (2R,3R,4R,5R,6S)-2,6-dimethyl-4-(3-nitrophenyl)piperidine-3,5-dicarboxylate, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wei, Y, Yu, Z, Zhao, Y. | | Deposit date: | 2022-12-02 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural bases of inhibitory mechanism of Ca V 1.2 channel inhibitors.
Nat Commun, 15, 2024
|
|
8HMA
 
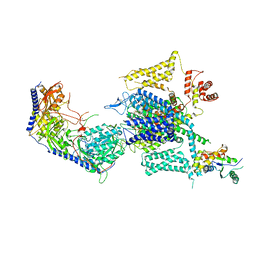 | | Cryo-EM structure of human high-voltage activated L-type calcium channel CaV1.2 in complex with tetrandrine (TET) | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2-acetamido-2-deoxy-beta-D-glucopyranose, 6,6',7,12-tetramethoxy-2,2'-dimethyl-1beta-3,4-didehydroberbaman, ... | | Authors: | Wei, Y, Yu, Z, Zhao, Y. | | Deposit date: | 2022-12-02 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural bases of inhibitory mechanism of Ca V 1.2 channel inhibitors.
Nat Commun, 15, 2024
|
|
2MXP
 
 | |
6JLH
 
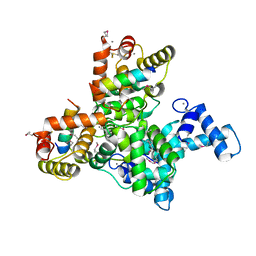 | | Structure of SCGN in complex with a Snap25 peptide | | Descriptor: | CALCIUM ION, CHLORIDE ION, Secretagogin, ... | | Authors: | Qin, J, Sun, Q, Jia, D. | | Deposit date: | 2019-03-05 | | Release date: | 2020-03-11 | | Last modified: | 2020-04-08 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural and mechanistic insights into secretagogin-mediated exocytosis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
8WC7
 
 | | Cryo-EM structure of the ZH8667-bound mTAAR1-Gs complex | | Descriptor: | 2-[4-(3-fluorophenyl)phenyl]ethanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC3
 
 | | Cryo-EM structure of the SEP363856-bound mTAAR1-Gs complex | | Descriptor: | 1-[(7~{S})-5,7-dihydro-4~{H}-thieno[2,3-c]pyran-7-yl]-~{N}-methyl-methanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WCA
 
 | | Cryo-EM structure of the PEA-bound hTAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC4
 
 | | Cryo-EM structure of the ZH8651-bound mTAAR1-Gs complex | | Descriptor: | 2-(4-bromophenyl)ethanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC9
 
 | | Cryo-EM structure of the ZH8651-bound mTAAR1-Gq complex | | Descriptor: | 2-(4-bromophenyl)ethanamine, Engineered G-alpha-q subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WCB
 
 | | Cryo-EM structure of the CHA-bound mTAAR1-Gq complex | | Descriptor: | CYCLOHEXYLAMMONIUM ION, Engineered G-alpha-q subunit, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC5
 
 | | Cryo-EM structure of the TMA-bound mTAAR1-Gs complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC8
 
 | | Cryo-EM structure of the ZH8651-bound hTAAR1-Gs complex | | Descriptor: | 2-(4-bromophenyl)ethanamine, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WC6
 
 | | Cryo-EM structure of the PEA-bound mTAAR1-Gs complex | | Descriptor: | 2-PHENYLETHYLAMINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
8WCC
 
 | | Cryo-EM structure of the CHA-bound mTAAR1 complex | | Descriptor: | CYCLOHEXYLAMMONIUM ION, Trace amine-associated receptor 1 | | Authors: | Rong, N.K, Guo, L.L, Zhang, M.H, Li, Q, Yang, F, Sun, J.P. | | Deposit date: | 2023-09-11 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural and signaling mechanisms of TAAR1 enabled preferential agonist design.
Cell, 186, 2023
|
|
6JEA
 
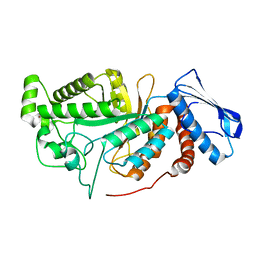 | | crystal structure of a beta-N-acetylhexosaminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-N-acetylhexosaminidase, ZINC ION | | Authors: | Chen, X, Wang, J.C, Liu, M.J, Yang, W.Y, Wang, Y.Z, Tang, R.P, Zhang, M. | | Deposit date: | 2019-02-04 | | Release date: | 2019-03-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.275 Å) | | Cite: | Crystallographic evidence for substrate-assisted catalysis of beta-N-acetylhexosaminidas from Akkermansia muciniphila.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
4RE8
 
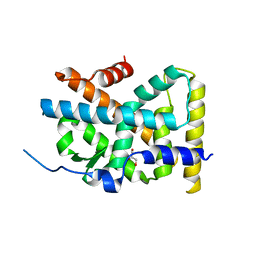 | | Crystal Structure of TR3 LBD in complex with Molecule 5 | | Descriptor: | 1-(3,4,5-trihydroxyphenyl)dodecan-1-one, GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Li, F.W, Cai, Q.X, Li, A.Z, Tian, X.Y, Weijia, W, Yuan, W, Hou, P.P, Wu, Q, Lin, T.W. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Induction of Autophagic Death in Cancer Cells by Agonizing TR3 and Attenuating Akt2 Activity
Chem.Biol., 22, 2015
|
|
5WB6
 
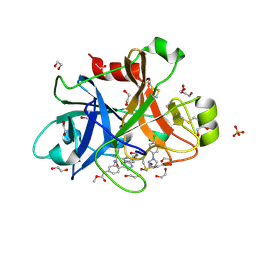 | | FACTOR XIA IN COMPLEX WITH THE INHIBITOR methyl [(11S)-11-({(2E)-3-[5-chloro-2-(1H-tetrazol-1-yl)phenyl]prop-2-enoyl}amino)-6-fluoro-2-oxo-1,3,4,10,11,13-hexahydro-2H-5,9:15,12-di(azeno)-1,13-benzodiazacycloheptadecin-18-yl]carbamate | | Descriptor: | 1,2-ETHANEDIOL, Coagulation factor XI, SULFATE ION, ... | | Authors: | Sheriff, S. | | Deposit date: | 2017-06-28 | | Release date: | 2017-08-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Macrocyclic factor XIa inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
7WOI
 
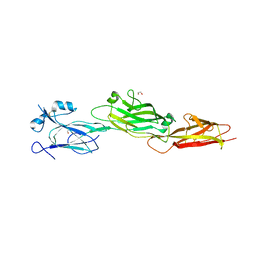 | | Structure of the shaft pilin Spa2 from Corynebacterium glutamicum | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Spa2 | | Authors: | Wu, Y.F, Wang, L.T, Huang, Y.Y, Zhong, C, Zhou, J. | | Deposit date: | 2022-01-21 | | Release date: | 2023-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Accelerating the design of pili-enabled living materials using an integrative technological workflow.
Nat.Chem.Biol., 2023
|
|
8WMI
 
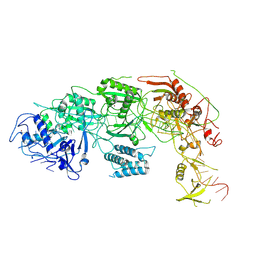 | |
8WM4
 
 | | Cryo-EM structure of DiCas7-11 in complex with crRNA | | Descriptor: | CRISPR-associated RAMP family protein, ZINC ION, crRNA (38-MER) | | Authors: | Ma, H.Y, Tang, X.D. | | Deposit date: | 2023-10-02 | | Release date: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | Structural basis of negative regulation of CRISPR-Cas7-11 by TPR-CHAT.
Signal Transduct Target Ther, 9, 2024
|
|
