6AKW
 
 | | Crystal structure of RNA dioxygenase bound with an inhibitor | | Descriptor: | 2-OXOGLUTARIC ACID, 2-[[2,6-bis(chloranyl)-4-(3,5-dimethyl-1,2-oxazol-4-yl)phenyl]amino]benzoic acid, Alpha-ketoglutarate-dependent dioxygenase FTO | | Authors: | Yang, C.-G, Huang, Y, Gan, J. | | Deposit date: | 2018-09-04 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia.
Cancer Cell, 35, 2019
|
|
6ILC
 
 | |
6ILF
 
 | |
6ILE
 
 | | CRYSTAL STRUCTURE OF A MUTANT PTAL-N*01:01 FOR 2.9 ANGSTROM, 52M 53D 54L DELETED | | Descriptor: | Beta-2-microglobulin, HEV-1, MHC class I antigen | | Authors: | Qu, Z.H, Zhang, N.Z, Xia, C. | | Deposit date: | 2018-10-17 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure and Peptidome of the Bat MHC Class I Molecule Reveal a Novel Mechanism Leading to High-Affinity Peptide Binding.
J Immunol., 202, 2019
|
|
6LJS
 
 | | Crystal structure of human FABP4 in complex with a novel inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2-phenylphenyl)amino]benzoic acid, Fatty acid-binding protein, ... | | Authors: | Su, H.X, Zhang, X.L, Li, M.J, Xu, Y.C. | | Deposit date: | 2019-12-17 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Exploration of Fragment Binding Poses Leading to Efficient Discovery of Highly Potent and Orally Effective Inhibitors of FABP4 for Anti-inflammation.
J.Med.Chem., 63, 2020
|
|
6ILG
 
 | |
6IRL
 
 | |
6KX9
 
 | |
5BWA
 
 | | Crystal structure of ODC-PLP-AZ1 ternary complex | | Descriptor: | Ornithine decarboxylase, Ornithine decarboxylase antizyme 1, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Wu, D.H. | | Deposit date: | 2015-06-07 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis of Ornithine Decarboxylase inactivation and accelerated degradation by polyamine sensor Antizyme1
Sci Rep, 5, 2015
|
|
5EB9
 
 | | Crystal Structure Of Chicken CD8aa Homodimer | | Descriptor: | CD8 alpha chain | | Authors: | Liu, Y.J, Qi, J.X, Xia, C. | | Deposit date: | 2015-10-18 | | Release date: | 2016-09-14 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | The structural basis of chicken, swine and bovine CD8 alpha alpha dimers provides insight into the co-evolution with MHC I in endotherm species.
Sci Rep, 6, 2016
|
|
8DW2
 
 | | Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV-2 DARPin,SR22, and two antibody Fabs, S309 and CR3022 | | Descriptor: | Antibody CR3022 heavy chain, Antibody CR3022 light chain, Antibody S309 heavy chain, ... | | Authors: | Kwon, Y.D, Gorman, J, Kwong, P.D. | | Deposit date: | 2022-07-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.11 Å) | | Cite: | A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins.
Nat.Chem.Biol., 19, 2023
|
|
8DW3
 
 | | Cryo-EM structure of SARS-CoV-2 RBD in complex with anti-SARS-CoV-2 DARPin,SR16m, and two antibody Fabs, S309 and CR3022 | | Descriptor: | Anti-SARS-CoV-2 DARPin SR16m, Antibody S309 light chain, Spike protein S1, ... | | Authors: | Kwon, Y.D, Gorman, J, Kwong, P.D. | | Deposit date: | 2022-07-30 | | Release date: | 2022-12-07 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.26 Å) | | Cite: | A potent and broad neutralization of SARS-CoV-2 variants of concern by DARPins.
Nat.Chem.Biol., 19, 2023
|
|
7KXT
 
 | | Crystal structure of human EED | | Descriptor: | 1-[(4-fluorophenyl)methyl]-N-{1-[2-(4-methoxyphenyl)ethyl]piperidin-4-yl}-1H-benzimidazol-2-amine, Polycomb protein EED, UNKNOWN ATOM OR ION | | Authors: | Zhu, L, Dong, A, Du, D, Liu, Y, Luo, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-12-04 | | Release date: | 2021-02-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Guided Development of Small-Molecule PRC2 Inhibitors Targeting EZH2-EED Interaction.
J.Med.Chem., 64, 2021
|
|
4ZY8
 
 | |
3S2A
 
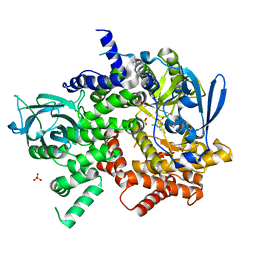 | | Crystal structure of PI3K-gamma in complex with a quinoline inhibitor | | Descriptor: | N-{2-chloro-5-[4-(morpholin-4-yl)quinolin-6-yl]pyridin-3-yl}-4-fluorobenzenesulfonamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Phospshoinositide 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Dual Inhibitors: Discovery and Structure-Activity Relationships of a Series of Quinoline and Quinoxaline Derivatives.
J.Med.Chem., 54, 2011
|
|
6A0Q
 
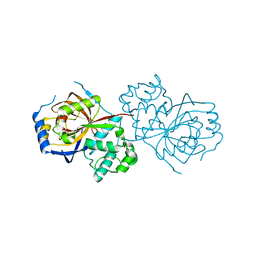 | | The crystal structure of Lpg2622_E64 complex | | Descriptor: | Lpg2622, N-[N-[1-HYDROXYCARBOXYETHYL-CARBONYL]LEUCYLAMINO-BUTYL]-GUANIDINE | | Authors: | Gong, X, Ge, H. | | Deposit date: | 2018-06-06 | | Release date: | 2018-09-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of the hypothetical protein Lpg2622, a new member of the C1 family peptidases from Legionella pneumophila
FEBS Lett., 592, 2018
|
|
6A0N
 
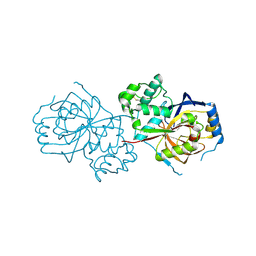 | | The crystal structure of apo-Lpg2622 | | Descriptor: | Lpg2622 | | Authors: | Gong, X, Ge, H. | | Deposit date: | 2018-06-05 | | Release date: | 2018-09-12 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of the hypothetical protein Lpg2622, a new member of the C1 family peptidases from Legionella pneumophila
FEBS Lett., 592, 2018
|
|
3QK0
 
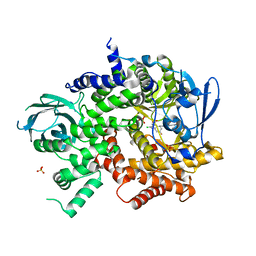 | | Crystal structure of PI3K-gamma in complex with benzothiazole 82 | | Descriptor: | N-[6-(6-chloro-5-{[(4-fluorophenyl)sulfonyl]amino}pyridin-3-yl)-1,3-benzothiazol-2-yl]acetamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2011-01-31 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Discovery and Optimization of a Series of Benzothiazole Phosphoinositide 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Dual Inhibitors.
J.Med.Chem., 54, 2011
|
|
4RBW
 
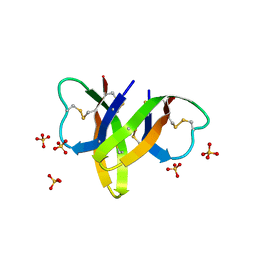 | | Crystal structure of human alpha-defensin 5, HD5 (Thr7Arg Glu21Arg mutant) | | Descriptor: | CHLORIDE ION, Defensin-5, SULFATE ION | | Authors: | Pazgier, M, Gohain, N, Tolbert, W.D. | | Deposit date: | 2014-09-13 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Design of a potent antibiotic peptide based on the active region of human defensin 5.
J.Med.Chem., 58, 2015
|
|
3QJZ
 
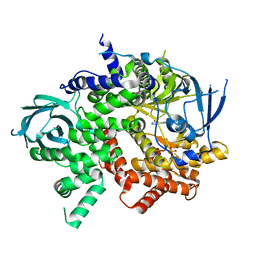 | | Crystal structure of PI3K-gamma in complex with benzothiazole 1 | | Descriptor: | N-{6-[2-(methylsulfanyl)pyrimidin-4-yl]-1,3-benzothiazol-2-yl}acetamide, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2011-01-31 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Discovery and Optimization of a Series of Benzothiazole Phosphoinositide 3-Kinase (PI3K)/Mammalian Target of Rapamycin (mTOR) Dual Inhibitors.
J.Med.Chem., 54, 2011
|
|
4FLH
 
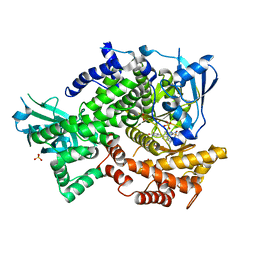 | | Crystal structure of human PI3K-gamma in complex with AMG511 | | Descriptor: | 4-(2-[(5-fluoro-6-methoxypyridin-3-yl)amino]-5-{(1R)-1-[4-(methylsulfonyl)piperazin-1-yl]ethyl}pyridin-3-yl)-6-methyl-1,3,5-triazin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2012-06-14 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Selective Class I Phosphoinositide 3-Kinase Inhibitors: Optimization of a Series of Pyridyltriazines Leading to the Identification of a Clinical Candidate, AMG 511.
J.Med.Chem., 55, 2012
|
|
6KWN
 
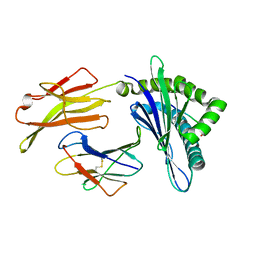 | | Crystal structure of pSLA-1*1301(F99Y) complex with S-OIV-derived epitope NSDTVGWSW | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, peptide | | Authors: | Wei, X.H, Wang, S, Zhang, N.Z, Xia, C. | | Deposit date: | 2019-09-07 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Peptidomes and Structures Illustrate Two Distinguishing Mechanisms of Alternating the Peptide Plasticity Caused by Swine MHC Class I Micropolymorphism.
Front Immunol, 12, 2021
|
|
6KWL
 
 | | Crystal structure of pSLA-1*0401(R156A) complex with FMDV-derived epitope MTAHITVPY | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, peptide | | Authors: | Wei, X.H, Wang, S, Zhang, N.Z, Xia, C. | | Deposit date: | 2019-09-07 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Peptidomes and Structures Illustrate Two Distinguishing Mechanisms of Alternating the Peptide Plasticity Caused by Swine MHC Class I Micropolymorphism.
Front Immunol, 12, 2021
|
|
6KWO
 
 | | Crystal structure of pSLA-1*1301 complex with mutant epitope ESDTVGWSW | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, peptide | | Authors: | Wei, X.H, Wang, S, Zhang, N.Z, Xia, C. | | Deposit date: | 2019-09-07 | | Release date: | 2020-09-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Peptidomes and Structures Illustrate Two Distinguishing Mechanisms of Alternating the Peptide Plasticity Caused by Swine MHC Class I Micropolymorphism.
Front Immunol, 12, 2021
|
|
4F1S
 
 | | Crystal structure of human PI3K-gamma in complex with a pyridyl-triazine-sulfonamide inhibitor | | Descriptor: | N-(5-{[3-(4-amino-6-methyl-1,3,5-triazin-2-yl)-5-(tetrahydro-2H-pyran-4-yl)pyridin-2-yl]amino}-2-chloropyridin-3-yl)methanesulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2012-05-07 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis and structure-activity relationships of dual PI3K/mTOR inhibitors based on a 4-amino-6-methyl-1,3,5-triazine sulfonamide scaffold.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
