5WXB
 
 | |
4LVA
 
 | | Fragment-based Identification of Amides Derived From trans-2-(Pyridin-3-yl)cyclopropanecarboxylic Acid as Potent Inhibitors of Human Nicotinamide Phosphoribosyltransferase (NAMPT) | | Descriptor: | 1,2-ETHANEDIOL, N-(4-{[4-(pyrrolidin-1-yl)piperidin-1-yl]sulfonyl}benzyl)-2H-pyrido[4,3-e][1,2,4]thiadiazin-3-amine 1,1-dioxide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Giannetti, A.M, Zheng, X, Skelton, N, Wang, W, Bravo, B, Feng, Y, Gunzner-Toste, J, Ho, Y, Hua, R, Wang, C, Zhao, Q, Liederer, B.M, Liu, Y, O'Brien, T, Oeh, J, Sampath, D, Shen, Y, Wang, L, Wu, H, Xiao, Y, Yuen, P, Zak, M, Zhao, G, Dragovich, P.S. | | Deposit date: | 2013-07-26 | | Release date: | 2013-09-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification of amides derived from 1H-pyrazolo[3,4-b]pyridine-5-carboxylic acid as potent inhibitors of human nicotinamide phosphoribosyltransferase (NAMPT).
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4OJR
 
 | | Crystal Structure of the HIV-1 Integrase catalytic domain with GSK1264 | | Descriptor: | (2S)-tert-butoxy[4-(8-fluoro-5-methyl-3,4-dihydro-2H-chromen-6-yl)-2-methyl-1-oxo-1,2-dihydroisoquinolin-3-yl]ethanoic acid, CACODYLATE ION, HIV-1 Integrase, ... | | Authors: | Gupta, K, Brady, T, Dyer, B, Hwang, Y, Male, F, Nolte, R.T, Wang, L, Velthuisen, E, Jeffrey, J, Van Duyne, G, Bushman, F.D. | | Deposit date: | 2014-01-21 | | Release date: | 2014-06-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Allosteric Inhibition of Human Immunodeficiency Virus Integrase: LATE BLOCK DURING VIRAL REPLICATION AND ABNORMAL MULTIMERIZATION INVOLVING SPECIFIC PROTEIN DOMAINS.
J.Biol.Chem., 289, 2014
|
|
4LT9
 
 | | Crystal Structure of Engineered Protein, Northeast Structural Genomics Consortium Target OR404 | | Descriptor: | Engineered Protein OR404 | | Authors: | Vorobiev, S, Su, M, Bjelic, S, Kipnis, Y, Wang, L, Sahdev, S, Xiao, R, Kogan, S, Maglaqui, M, Baker, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-07-23 | | Release date: | 2013-08-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of Engineered Protein OR404.
To be Published
|
|
3JCM
 
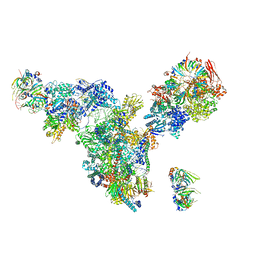 | | Cryo-EM structure of the spliceosomal U4/U6.U5 tri-snRNP | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, GUANOSINE-5'-TRIPHOSPHATE, N,N,7-trimethylguanosine 5'-(trihydrogen diphosphate), ... | | Authors: | Wan, R, Yan, C, Bai, R, Wang, L, Huang, M, Wong, C.C, Shi, Y. | | Deposit date: | 2015-12-23 | | Release date: | 2016-02-24 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The 3.8 angstrom structure of the U4/U6.U5 tri-snRNP: Insights into spliceosome assembly and catalysis
Science, 351, 2016
|
|
5W0Z
 
 | |
5W0R
 
 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with cacodylic acid | | Descriptor: | CACODYLATE ION, CALCIUM ION, MBP fused activation-induced cytidine deaminase, ... | | Authors: | Qiao, Q, Wang, L, Wu, H. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
5W0U
 
 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with dCMP | | Descriptor: | 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*TP*GP*AP*AP*C)-3'), ... | | Authors: | Qiao, Q, Wang, L, Wu, H. | | Deposit date: | 2017-05-31 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
5W1C
 
 | | Crystal structure of MBP fused activation-induced cytidine deaminase (AID) in complex with cytidine | | Descriptor: | 4-AMINO-1-BETA-D-RIBOFURANOSYL-2(1H)-PYRIMIDINONE, CALCIUM ION, DNA (5'-D(*CP*TP*GP*GP*CP*CP*TP*TP*GP*AP*AP*C)-3'), ... | | Authors: | Qiao, Q, Wang, L, Wu, H. | | Deposit date: | 2017-06-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | AID Recognizes Structured DNA for Class Switch Recombination.
Mol. Cell, 67, 2017
|
|
4WQN
 
 | | Crystal structure of N6-methyladenosine RNA reader YTHDF2 | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, YTH domain-containing family protein 2 | | Authors: | Zhu, T, Roundtree, I.A, Wang, P, Wang, X, Wang, L, Sun, C, Tian, Y, Li, J, He, C, Xu, Y. | | Deposit date: | 2014-10-22 | | Release date: | 2014-11-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.121 Å) | | Cite: | Crystal structure of the YTH domain of YTHDF2 reveals mechanism for recognition of N6-methyladenosine.
Cell Res., 24, 2014
|
|
5WVZ
 
 | | The crystal structure of Cren7 mutant L28F in complex with dsDNA | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*CP*GP*AP*TP*CP*GP*C)-3') | | Authors: | Zhang, Z.F, Zhao, M.H, Wang, L, Chen, Y.Y, Dong, Y.H, Gong, Y, Huang, L. | | Deposit date: | 2016-12-30 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Roles of Leu28 side chain intercalation in the interaction between Cren7 and DNA
Biochem. J., 474, 2017
|
|
5WVY
 
 | | The crystal structure of Cren7 mutant L28V in complex with dsDNA | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*GP*AP*TP*CP*AP*C)-3') | | Authors: | Zhang, Z.F, Zhao, M.H, Wang, L, Chen, Y.Y, Dong, Y.H, Gong, Y, Huang, L. | | Deposit date: | 2016-12-29 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Roles of Leu28 side chain intercalation in the interaction between Cren7 and DNA
Biochem. J., 474, 2017
|
|
5WVW
 
 | | The crystal structure of Cren7 mutant L28A in complex with dsDNA | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*GP*AP*TP*CP*AP*C)-3') | | Authors: | Zhang, Z.F, Zhao, M.H, Wang, L, Chen, Y.Y, Dong, Y.H, Gong, Y, Huang, L. | | Deposit date: | 2016-12-29 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Roles of Leu28 side chain intercalation in the interaction between Cren7 and DNA
Biochem. J., 474, 2017
|
|
7WT9
 
 | | SARS-CoV-2 Omicron variant spike RBD in complex with Fab 9A8 | | Descriptor: | Heavy chain of Fab 9A8, Light chain of Fab 9A8, Spike glycoprotein | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-02-04 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | A broader neutralizing antibody against all the current VOCs and VOIs targets unique epitope of SARS-CoV-2 RBD.
Cell Discov, 8, 2022
|
|
7WT7
 
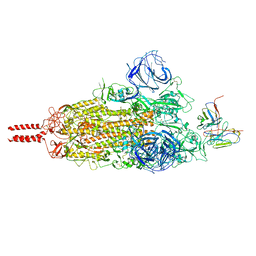 | | SARS-CoV-2 Omicron variant spike in complex with Fab 9A8 (State 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 9A8, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-02-04 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A broader neutralizing antibody against all the current VOCs and VOIs targets unique epitope of SARS-CoV-2 RBD.
Cell Discov, 8, 2022
|
|
7WT8
 
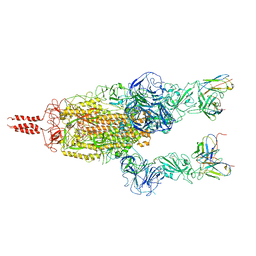 | | SARS-CoV-2 Omicron variant spike in complex with Fab 9A8 (State 2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 9A8, ... | | Authors: | Wang, X, Wang, L. | | Deposit date: | 2022-02-04 | | Release date: | 2023-06-07 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A broader neutralizing antibody against all the current VOCs and VOIs targets unique epitope of SARS-CoV-2 RBD.
Cell Discov, 8, 2022
|
|
5WWC
 
 | | The crystal structure of Cren7 mutant L28M in complex with dsDNA | | Descriptor: | Chromatin protein Cren7, DNA (5'-D(*GP*TP*AP*AP*TP*TP*AP*C)-3') | | Authors: | Zhang, Z.F, Zhao, M.H, Wang, L, Chen, Y.Y, Dong, Y.H, Gong, Y, Huang, L. | | Deposit date: | 2016-12-31 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Roles of Leu28 side chain intercalation in the interaction between Cren7 and DNA
Biochem. J., 474, 2017
|
|
4M00
 
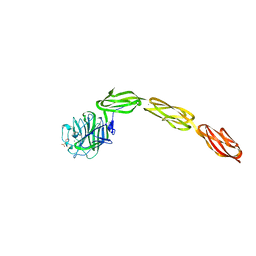 | | Crystal structure of the ligand binding region of staphylococcal adhesion SraP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, Serine-rich adhesin for platelets, ... | | Authors: | Yang, Y.H, Jiang, Y.L, Zhang, J, Wang, L, Chen, Y, Zhou, C.Z. | | Deposit date: | 2013-08-01 | | Release date: | 2014-06-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Insights into SraP-Mediated Staphylococcus aureus Adhesion to Host Cells
Plos Pathog., 10, 2014
|
|
8ZDW
 
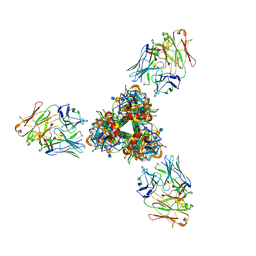 | | The cryoEM structure of H5N1 HA split from symmetric filament in conformation A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Anti-H5N1 hemagglutinin monoclonal anitbody H5M9 heavy chain, ... | | Authors: | Li, R, Gao, J, Wang, L, Gui, M, Xiang, Y. | | Deposit date: | 2024-05-03 | | Release date: | 2024-09-25 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Multivalent interactions between fully glycosylated influenza virus hemagglutinins mediated by glycans at distinct N-glycosylation sites
To Be Published
|
|
8ZDV
 
 | | The cryoEM structure of H5N8 HA in an auto inhibited state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Li, R, Gao, J, Wang, L, Gui, M, Xiang, Y. | | Deposit date: | 2024-05-03 | | Release date: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Multivalent interactions between fully glycosylated influenza virus hemagglutinins mediated by glycans at distinct N-glycosylation sites
To Be Published
|
|
4WGI
 
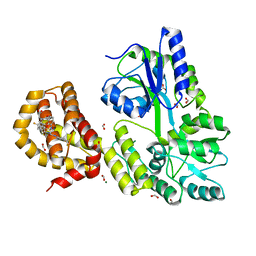 | | A Single Diastereomer of a Macrolactam Core Binds Specifically to Myeloid Cell Leukemia 1 (MCL1) | | Descriptor: | (2S)-2-[(2S,3R)-10-{[(4-fluorophenyl)sulfonyl]amino}-3-methyl-2-[(methyl{[4-(trifluoromethyl)phenyl]carbamoyl}amino)methyl]-6-oxo-3,4-dihydro-2H-1,5-benzoxazocin-5(6H)-yl]propanoic acid, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Fairman, J.W, Fang, C, D'Souza, B, Fulroth, B, Leed, A, McCarren, P, Wang, L, Wang, Y, Kaushik, V, Palmer, M, Wei, G, Golub, T.R, Hubbard, B.K, Serrano-Wu, M.H. | | Deposit date: | 2014-09-18 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Single Diastereomer of a Macrolactam Core Binds Specifically to Myeloid Cell Leukemia 1 (MCL1).
Acs Med.Chem.Lett., 5, 2014
|
|
7Y42
 
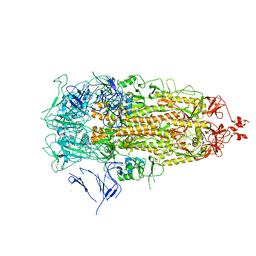 | |
4U7T
 
 | | Crystal structure of DNMT3A-DNMT3L in complex with histone H3 | | Descriptor: | DNA (cytosine-5)-methyltransferase 3-like, DNA (cytosine-5)-methyltransferase 3A, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Guo, X, Wang, L, Yin, X, Li, J, Xiao, J, He, S, Wang, J, Xu, Y. | | Deposit date: | 2014-07-31 | | Release date: | 2014-11-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insight into autoinhibition and histone H3-induced activation of DNMT3A
Nature, 517, 2015
|
|
8YJY
 
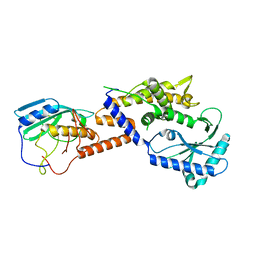 | |
4R8X
 
 | | Crystal structure of a uricase from Bacillus fastidious | | Descriptor: | Uricase | | Authors: | Feng, J, Wang, L, Liu, H.B, Liu, L, Liao, F. | | Deposit date: | 2014-09-03 | | Release date: | 2015-05-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Crystal structure of Bacillus fastidious uricase reveals an unexpected folding of the C-terminus residues crucial for thermostability under physiological conditions.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
