8BH5
 
 | | SARS-CoV-2 BA.2.12.1 RBD in complex with Beta-27 Fab and C1 nanobody | | Descriptor: | Beta-27 heavy chain, Beta-27 light chain, GLYCEROL, ... | | Authors: | Huo, J, Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2022-10-29 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Humoral responses against SARS-CoV-2 Omicron BA.2.11, BA.2.12.1 and BA.2.13 from vaccine and BA.1 serum.
Cell Discov, 8, 2022
|
|
4E2T
 
 | | Crystal Structures of RadA intein from Pyrococcus horikoshii | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Pho radA intein | | Authors: | Oeemig, J.S, Zhou, D, Kajander, T, Wlodawer, A, Iwai, H. | | Deposit date: | 2012-03-09 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | NMR and Crystal Structures of the Pyrococcus horikoshii RadA Intein Guide a Strategy for Engineering a Highly Efficient and Promiscuous Intein.
J.Mol.Biol., 421, 2012
|
|
4E2U
 
 | | Crystal Structures of RadAmin intein from Pyrococcus horikoshii | | Descriptor: | Pho radA intein | | Authors: | Oeemig, J.S, Zhou, D, Kajander, T, Wlodawer, A, Iwai, H. | | Deposit date: | 2012-03-09 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.582 Å) | | Cite: | NMR and Crystal Structures of the Pyrococcus horikoshii RadA Intein Guide a Strategy for Engineering a Highly Efficient and Promiscuous Intein.
J.Mol.Biol., 421, 2012
|
|
4PKM
 
 | | Crystal Structure of Bacillus thuringiensis Cry51Aa1 Protoxin at 1.65 Angstroms Resolution | | Descriptor: | Cry51Aa1, GLYCEROL, GLYCINE, ... | | Authors: | Xu, C, Chinte, U, Chen, L, Yao, Q, Zhou, D, Meng, Y, Li, L, Rose, J, Bi, L.J, Yu, Z, Sun, M, Wang, B.C. | | Deposit date: | 2014-05-15 | | Release date: | 2015-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of Cry51Aa1: A potential novel insecticidal aerolysin-type beta-pore-forming toxin from Bacillus thuringiensis.
Biochem.Biophys.Res.Commun., 462, 2015
|
|
8CIN
 
 | | BA.4/5-5 FAB IN COMPLEX WITH SARS-COV-2 BA.4 SPIKE GLYCOPROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BA.4/5-5 fab HEAVY CHAIN, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I, Fry, E.E. | | Deposit date: | 2023-02-10 | | Release date: | 2024-02-21 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The SARS-CoV-2 neutralizing antibody response to SD1 and its evasion by BA.2.86.
Nat Commun, 15, 2024
|
|
8R1C
 
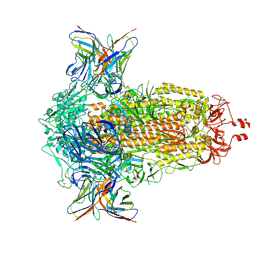 | | SD1-2 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SD1-2 fab heavy chain, SD1-2 fab light chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2023-11-01 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | The SARS-CoV-2 neutralizing antibody response to SD1 and its evasion by BA.2.86.
Nat Commun, 15, 2024
|
|
8R1D
 
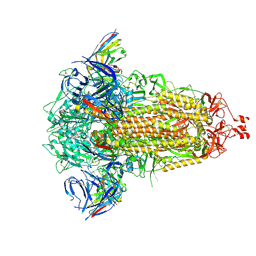 | | SD1-3 Fab in complex with SARS-CoV-2 BA.2.12.1 Spike Glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, SD1-3 Fab Heavy Chain, SD1-3 Fab Light Chain, ... | | Authors: | Duyvesteyn, H.M.E, Ren, J, Stuart, D.I. | | Deposit date: | 2023-11-01 | | Release date: | 2024-03-13 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | The SARS-CoV-2 neutralizing antibody response to SD1 and its evasion by BA.2.86.
Nat Commun, 15, 2024
|
|
5Z1G
 
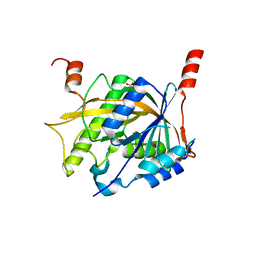 | | Structure of the Brx1 and Ebp2 complex | | Descriptor: | Ribosome biogenesis protein BRX1, SULFATE ION, rRNA-processing protein EBP2 | | Authors: | Zheng, S, Ye, K. | | Deposit date: | 2017-12-26 | | Release date: | 2018-04-11 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Cryo-EM structure of an early precursor of large ribosomal subunit reveals a half-assembled intermediate
Protein Cell, 10, 2019
|
|
8HJE
 
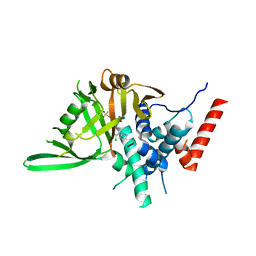 | | Vismodegib binds to the catalytical domain of human Ubiquitin-Specific Protease 28 | | Descriptor: | 2-chloranyl-~{N}-(4-chloranyl-3-pyridin-2-yl-phenyl)-4-methylsulfonyl-benzamide, Ubiquitin carboxyl-terminal hydrolase 28 | | Authors: | Shi, L, Wang, H, Xu, Z, Xiong, B, Zhang, N. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure-based discovery of potent USP28 inhibitors derived from Vismodegib.
Eur.J.Med.Chem., 254, 2023
|
|
4WBQ
 
 | | Crystal structure of the exonuclease domain of QIP (QDE-2 interacting protein) solved by native-SAD phasing. | | Descriptor: | CALCIUM ION, QDE-2-interacting protein | | Authors: | Boland, A, Weinert, T, Weichenrieder, O, Wang, M. | | Deposit date: | 2014-09-03 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4WAB
 
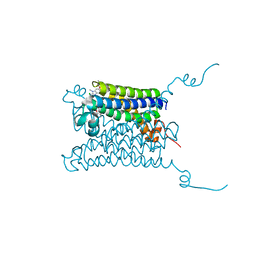 | | Crystal structure of mPGES1 solved by native-SAD phasing | | Descriptor: | 2-[[2,6-bis(chloranyl)-3-[(2,2-dimethylpropanoylamino)methyl]phenyl]amino]-1-methyl-6-(2-methyl-2-oxidanyl-propoxy)-N-[2,2,2-tris(fluoranyl)ethyl]benzimidazole-5-carboxamide, GLUTATHIONE, Prostaglandin E synthase,Leukotriene C4 synthase | | Authors: | Weinert, T, Li, D, Howe, N, Caffrey, M, Wang, M. | | Deposit date: | 2014-08-29 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.704 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4WBX
 
 | | Conserved hypothetical protein PF1771 from Pyrococcus furiosus solved by sulfur SAD using Swiss Light Source data | | Descriptor: | 2-keto acid:ferredoxin oxidoreductase subunit alpha | | Authors: | Weinert, T, Waltersperger, S, Olieric, V, Panepucci, E, Chen, L, Rose, J.P, Wang, M, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2014-09-04 | | Release date: | 2014-12-10 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
4WAU
 
 | | Crystal structure of CENP-M solved by native-SAD phasing | | Descriptor: | Centromere protein M | | Authors: | Weinert, T, Basilico, F, Cecatiello, V, Pasqualato, S, Wang, M. | | Deposit date: | 2014-09-01 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
8ZMR
 
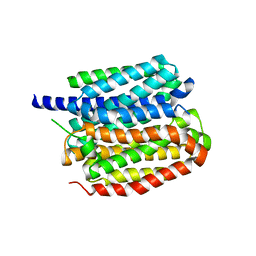 | | Vesamicol-bound VAChT | | Descriptor: | Maltose/maltodextrin-binding periplasmic protein,Vesicular acetylcholine transporter,DARPinoff7, vesamicol | | Authors: | Zhang, Z, Zhang, Y, Dai, F, Zhang, Y.X, Lee, C.-H. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into VAChT neurotransmitter recognition and inhibition.
Cell Res., 2024
|
|
8ZMS
 
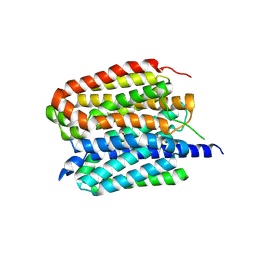 | | Acetylcholine-bound VAChT | | Descriptor: | ACETYLCHOLINE, Maltose/maltodextrin-binding periplasmic protein,Vesicular acetylcholine transporter,DARPinoff7 | | Authors: | Zhang, Z, Zhang, Y, Dai, F, Zhang, Y.X, Lee, C.-H. | | Deposit date: | 2024-05-23 | | Release date: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural insights into VAChT neurotransmitter recognition and inhibition.
Cell Res., 2024
|
|
6ZH9
 
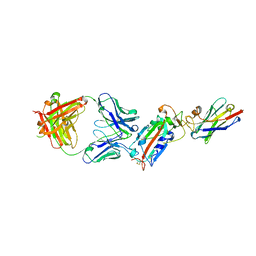 | | Ternary complex CR3022 H11-H4 and RBD (SARS-CoV-2) | | Descriptor: | CR3022 Light chain, CR3022 heavy, Nanobody H11-H4, ... | | Authors: | Naismith, J.H, Mikolajek, H, Le Bas, A. | | Deposit date: | 2020-06-21 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2.
Nat.Struct.Mol.Biol., 27, 2020
|
|
8R8K
 
 | |
3IK3
 
 | |
4NXL
 
 | | Dibenzothiophene monooxygenase (DszC) from Rhodococcus erythropolis | | Descriptor: | DszC | | Authors: | Zhang, L, Duan, X, Li, X, Rao, Z. | | Deposit date: | 2013-12-09 | | Release date: | 2014-07-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into the stabilization of active, tetrameric DszC by its C-terminus.
Proteins, 82, 2014
|
|
5KY9
 
 | |
5KY4
 
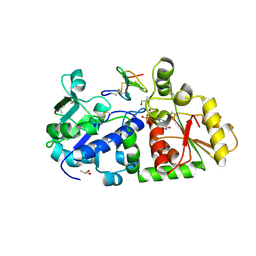 | | mouse POFUT1 in complex with mouse Notch1 EGF26 and GDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP-fucose protein O-fucosyltransferase 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-21 | | Release date: | 2017-05-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.472 Å) | | Cite: | Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1.
Nat. Chem. Biol., 13, 2017
|
|
5L0T
 
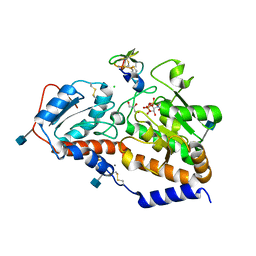 | | human POGLUT1 in complex with EGF(+) and UDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-28 | | Release date: | 2017-08-09 | | Last modified: | 2021-03-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural basis of Notch O-glucosylation and O-xylosylation by mammalian protein-O-glucosyltransferase 1 (POGLUT1).
Nat Commun, 8, 2017
|
|
5KY0
 
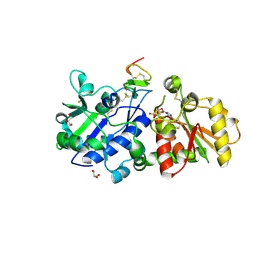 | | mouse POFUT1 in complex with mouse Notch1 EGF12(D464G) and GDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP-fucose protein O-fucosyltransferase 1, GLYCEROL, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-20 | | Release date: | 2017-05-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1.
Nat. Chem. Biol., 13, 2017
|
|
2QZA
 
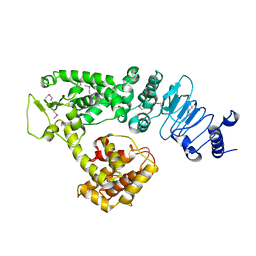 | | Crystal structure of Salmonella effector protein SopA | | Descriptor: | Secreted effector protein | | Authors: | Diao, J, Chen, J. | | Deposit date: | 2007-08-16 | | Release date: | 2007-12-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of SopA, a Salmonella effector protein mimicking a eukaryotic ubiquitin ligase.
Nat.Struct.Mol.Biol., 15, 2008
|
|
6YZ7
 
 | | H11-D4, SARS-CoV-2 RBD, CR3022 ternary complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Cr3022, Antibody light chain, ... | | Authors: | Naismith, J.H, Ren, J, Zhou, D, Zhao, Y, Stuart, D.I. | | Deposit date: | 2020-05-06 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural characterisation of a nanobody derived from a naive library that neutralises SARS-CoV-2
To Be Published, 2020
|
|
