6IBT
 
 | | Crystal structure of human alpha-galactosidase A in complex with alpha-galactose configured cyclophellitol aziridine ME737 | | Descriptor: | (1~{S},2~{S},3~{S},4~{S},5~{R},6~{S})-5-(hydroxymethyl)-7-azabicyclo[4.1.0]heptane-2,3,4-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Wu, L, Davies, G.J. | | Deposit date: | 2018-11-30 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Alpha-d-Gal-cyclophellitol cyclosulfamidate is a Michaelis complex analog that stabilizes therapeutic lysosomal alpha-galactosidase A in Fabry disease
Chem Sci, 2019
|
|
6IBK
 
 | | Crystal structure of human alpha-galactosidase A in complex with alpha-galactose configured cyclosulfamidate ME763 | | Descriptor: | (3~{a}~{R},4~{S},5~{S},6~{S},7~{R},7~{a}~{S})-7-(hydroxymethyl)-2,2-bis(oxidanylidene)-3~{a},4,5,6,7,7~{a}-hexahydro-3~{H}-benzo[d][1,2,3]oxathiazole-4,5,6-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rowland, R.J, Wu, L, Davies, G.J. | | Deposit date: | 2018-11-30 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Alpha-d-Gal-cyclophellitol cyclosulfamidate is a Michaelis complex analog that stabilizes therapeutic lysosomal alpha-galactosidase A in Fabry disease
Chem Sci, 2019
|
|
8A3H
 
 | | Cellobiose-derived imidazole complex of the endoglucanase cel5A from Bacillus agaradhaerens at 0.97 A resolution | | Descriptor: | (5R,6R,7R,8S)-7,8-dihydroxy-5-(hydroxymethyl)-5,6,7,8-tetrahydroimidazo[1,2-a]pyridin-6-yl beta-D-glucopyranoside, ACETATE ION, GLYCEROL, ... | | Authors: | Varrot, A, Schulein, M, Pipelier, M, Vasella, A, Davies, G.J. | | Deposit date: | 1999-01-20 | | Release date: | 2000-01-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Lateral Protonation of a Glycosidase Inhibitor. Structure of the Bacillus agaradhaerens Cel5A in Complex with a Cellobiose-Derived Imidazole at 0.97 A Resolution
J.Am.Chem.Soc., 121, 1999
|
|
8B2H
 
 | | Muramidase from Thermothielavioides terrestris, catalytic domain | | Descriptor: | 1,2-ETHANEDIOL, SH3b domain-containing protein, ZINC ION | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8B2F
 
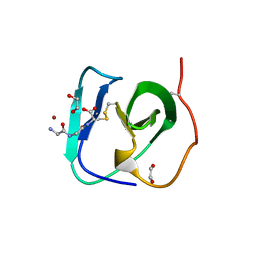 | | SH3-like cell wall binding domain of the GH24 family muramidase from Trichophaea saccata in complex with triglycine | | Descriptor: | 1,2-ETHANEDIOL, GLY-GLY-GLY, SH3-like cell wall binding domain-containing protein, ... | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.183 Å) | | Cite: | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8B2G
 
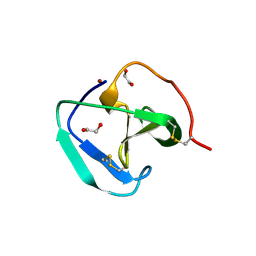 | | SH3-like domain from Penicillium virgatum muramidase | | Descriptor: | 1,2-ETHANEDIOL, SH3b domain-containing protein, ZINC ION | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8B2E
 
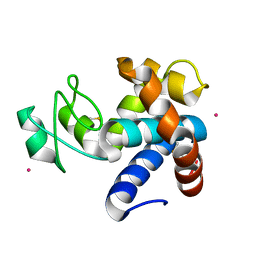 | | Muramidase from Kionochaeta sp natural catalytic core | | Descriptor: | CADMIUM ION, Muramidase | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | Deposit date: | 2022-09-13 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8B2S
 
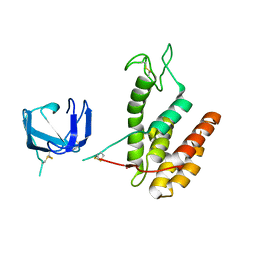 | | GH24 family muramidase from Trichophaea saccata with an SH3-like cell wall binding domain | | Descriptor: | GH24 family muramidase, POTASSIUM ION | | Authors: | Moroz, O.V, Blagova, E, Lebedev, A.A, Skov, L.K, Pache, R.A, Schnorr, K.M, Kiemer, L, Nymand-Grarup, S, Ming, L, Ye, L, Klausen, M, Cohn, M.T, Schmidt, E.G.W, Davies, G.J, Wilson, K.S. | | Deposit date: | 2022-09-14 | | Release date: | 2023-07-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Module walking using an SH3-like cell-wall-binding domain leads to a new GH184 family of muramidases.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
5C86
 
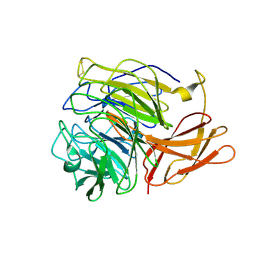 | | Novel fungal alcohol oxidase with catalytic diversity among the AA5 family, apo form | | Descriptor: | Kelch domain-containing protein | | Authors: | Urresti, S, Yin, D.T, LaFond, M, Derikvand, F, Berrin, G.J, Henrissat, B, Walton, P.H, Brumer, H, Davies, G.J. | | Deposit date: | 2015-06-25 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structure-function characterization reveals new catalytic diversity in the galactose oxidase and glyoxal oxidase family.
Nat Commun, 6, 2015
|
|
5C92
 
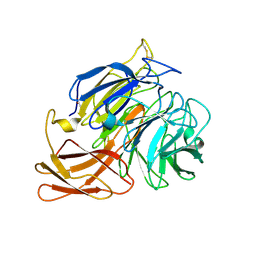 | | Novel fungal alcohol oxidase with catalytic diversity among the AA5 family, in complex with copper | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, COPPER (I) ION, ... | | Authors: | Urresti, S, Yin, D.T, LaFond, M, Derikvand, F, Berrin, G.J, Henrissat, B, Walton, P.H, Brumer, H, Davies, G.J. | | Deposit date: | 2015-06-26 | | Release date: | 2015-07-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-function characterization reveals new catalytic diversity in the galactose oxidase and glyoxal oxidase family.
Nat Commun, 6, 2015
|
|
2VI0
 
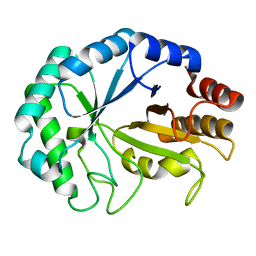 | | Lichenase CtLic26 in complex with a thio-oligosaccharide | | Descriptor: | 4-thio-beta-D-glucopyranose-(1-4)-methyl beta-D-glucopyranoside, Endoglucanase H, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Money, V.A, Ducros, V.M, Davies, G.J. | | Deposit date: | 2007-11-26 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Probing the beta-1,3:1,4 glucanase, CtLic26A, with a thio-oligosaccharide and enzyme variants.
Org. Biomol. Chem., 6, 2008
|
|
5IJU
 
 | | Structure of an AA10 Lytic Polysaccharide Monooxygenase from Bacillus amyloliquefaciens with Cu(II) bound | | Descriptor: | 1,2-ETHANEDIOL, BaAA10 Lytic Polysaccharide Monooxygenase, CALCIUM ION, ... | | Authors: | Gregory, R.C, Hemsworth, G.R, Turkenburg, J.P, Hart, S.J, Walton, P.H, Davies, G.J. | | Deposit date: | 2016-03-02 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Activity, stability and 3-D structure of the Cu(ii) form of a chitin-active lytic polysaccharide monooxygenase from Bacillus amyloliquefaciens.
Dalton Trans, 45, 2016
|
|
1OC6
 
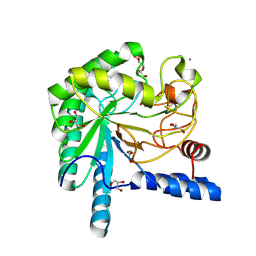 | | structure native of the D405N mutant of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS at 1.5 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CELLOBIOHYDROLASE II, ... | | Authors: | Varrot, A, Frandsen, T.P, Von Ossowski, I, Boyer, V, Driguez, H, Schulein, M, Davies, G.J. | | Deposit date: | 2003-02-06 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for Ligand Binding and Processivity in Cellobiohydrolase Cel6A from Humicola Insolens
Structure, 11, 2003
|
|
1OCQ
 
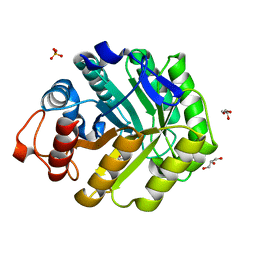 | | COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHEARANS AT 1.08 ANGSTROM RESOLUTION with cellobio-derived isofagomine | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, ENDOGLUCANASE 5A, GLYCEROL, ... | | Authors: | Varrot, A, Macdonald, J, Stick, R.V, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-02-09 | | Release date: | 2003-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Direct Observation of the Protonation State of an Imino Sugar Glycosidase Inhibitor Upon Binding
J.Am.Chem.Soc., 125, 2003
|
|
1NLR
 
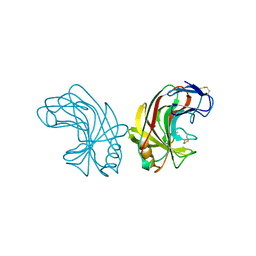 | | ENDO-1,4-BETA-GLUCANASE CELB2, CELLULASE, NATIVE STRUCTURE | | Descriptor: | ENDO-1,4-BETA-GLUCANASE | | Authors: | Sulzenbacher, G, Dupont, C, Davies, G.J. | | Deposit date: | 1997-10-27 | | Release date: | 1998-11-25 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Streptomyces lividans family 12 endoglucanase: construction of the catalytic cre, expression, and X-ray structure at 1.75 A resolution.
Biochemistry, 36, 1997
|
|
1OD3
 
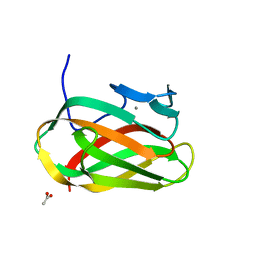 | | Structure of CSCBM6-3 From Clostridium stercorarium in complex with laminaribiose | | Descriptor: | ACETIC ACID, CALCIUM ION, PUTATIVE XYLANASE, ... | | Authors: | Boraston, A.B, Notenboom, V, Warren, R.A.J, Kilburn, D.G, Rose, D.R, Davies, G.J. | | Deposit date: | 2003-02-12 | | Release date: | 2003-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure and Ligand Binding of Carbohydrate-Binding Module Cscbm6-3 Reveals Similarities with Fucose-Specific Lectins and Galactose-Binding Domains
J.Mol.Biol., 327, 2003
|
|
1OCJ
 
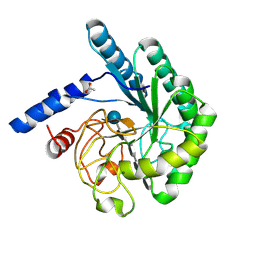 | | Mutant D416A of the CELLOBIOHYDROLASE CEL6A FROM HUMICOLA INSOLENS in complex with a THIOPENTASACCHARIDE at 1.3 angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CELLOBIOHYDROLASE II, ... | | Authors: | Varrot, A, Frandsen, T.P, Von Ossowski, I, Boyer, V, Driguez, H, Schulein, M, Davies, G.J. | | Deposit date: | 2003-02-07 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Basis for Ligand Binding and Processivity in Cellobiohydrolase Cel6A from Humicola Insolens
Structure, 11, 2003
|
|
1ODZ
 
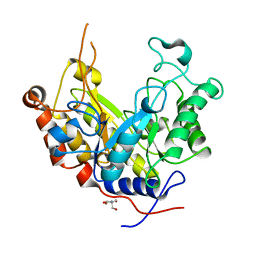 | | Expansion of the glycosynthase repertoire to produce defined manno-oligosaccharides | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Mannan endo-1,4-beta-mannosidase, SODIUM ION, ... | | Authors: | Jahn, M, Stoll, D, Warren, R.A.J, Szabo, L, Singh, P, Gilbert, H.J, Ducros, V.M.A, Davies, G.J, Withers, S.G. | | Deposit date: | 2003-03-17 | | Release date: | 2003-07-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Expansion of the Glycosynthase Repertoire to Produce Defined Manno-Oligosaccharides
Chem.Commun.(Camb.), 12, 2003
|
|
2IYF
 
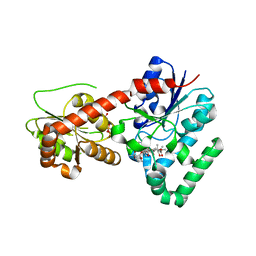 | | The crystal structure of macrolide glycosyltransferases: A blueprint for antibiotic engineering | | Descriptor: | ERYTHROMYCIN A, MAGNESIUM ION, OLEANDOMYCIN GLYCOSYLTRANSFERASE, ... | | Authors: | Bolam, D.N, Roberts, S.M, Proctor, M.R, Turkenburg, J.P, Dodson, E.J, Martinez-Fleites, C, Yang, M, Davis, B.G, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2006-07-17 | | Release date: | 2007-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Two Macrolide Glycosyltransferases Provides a Blueprint for Host Cell Antibiotic Immunity.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2IYA
 
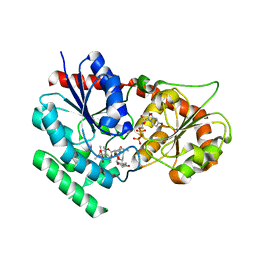 | | The crystal structure of macrolide glycosyltransferases: A blueprint for antibiotic engineering | | Descriptor: | (3S,5R,6S,7R,8R,11R,12S,13R,14S,15S)-6-HYDROXY-5,7,8,11,13,15-HEXAMETHYL-4,10-DIOXO-14-{[3,4,6-TRIDEOXY-3-(DIMETHYLAMINO)-BETA-D-XYLO-HEXOPYRANOSYL]OXY}-1,9-DIOXASPIRO[2.13]HEXADEC-12-YL 2,6-DIDEOXY-3-O-METHYL-ALPHA-L-ARABINO-HEXOPYRANOSIDE, OLEANDOMYCIN GLYCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Bolam, D.N, Roberts, S.M, Proctor, M.R, Turkenburg, J.P, Dodson, E.J, Martinez-Fleites, C, Yang, M, Davis, B.G, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2006-07-13 | | Release date: | 2007-03-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Crystal Structure of Two Macrolide Glycosyltransferases Provides a Blueprint for Host Cell Antibiotic Immunity.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
6ZFA
 
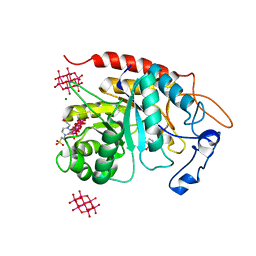 | | Structure of the catalytic domain of human endo-alpha-mannosidase MANEA in complex with GlcIFG, alpha-1,2-mannobiose and hexatungstotellurate(VI) TEW | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, 6-tungstotellurate(VI), ... | | Authors: | Sobala, L.F, Fernandes, P.Z, Hakki, Z, Thompson, A.J, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-16 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZFN
 
 | | Structure of an inactive E404Q variant of the catalytic domain of human endo-alpha-mannosidase MANEA in complex with 1-methyl alpha-1,2-mannobiose | | Descriptor: | Glycoprotein endo-alpha-1,2-mannosidase, SULFATE ION, alpha-D-mannopyranose-(1-2)-methyl alpha-D-mannopyranoside | | Authors: | Sobala, L.F, Fernandes, P.Z, Hakki, Z, Thompson, A.J, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZDC
 
 | | Structure of the catalytic domain of human endo-alpha-mannosidase MANEA in complex with nickel | | Descriptor: | Glycoprotein endo-alpha-1,2-mannosidase, NICKEL (II) ION | | Authors: | Sobala, L.F, Fernandes, P.Z, Hakki, Z, Thompson, A.J, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZFQ
 
 | | Structure of the catalytic domain of human endo-alpha-mannosidase MANEA in complex with bis-tris | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Glycoprotein endo-alpha-1,2-mannosidase | | Authors: | Sobala, L.F, Fernandes, P.Z, Hakki, Z, Thompson, A.J, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-17 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZDF
 
 | | Structure of the catalytic domain of human endo-alpha-mannosidase MANEA in complex with HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Glycoprotein endo-alpha-1,2-mannosidase | | Authors: | Sobala, L.F, Fernandes, P.Z, Hakki, Z, Thompson, A.J, Howe, J.D, Hill, M, Zitzmann, N, Davies, S, Stamataki, Z, Butters, T.D, Alonzi, D.S, Williams, S.J, Davies, G.J. | | Deposit date: | 2020-06-14 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of human endo-alpha-1,2-mannosidase (MANEA), an antiviral host-glycosylation target.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
