7JRC
 
 | | Phospholipase D engineered mutant in complex with phosphate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Vrielink, A, Samantha, A. | | Deposit date: | 2020-08-12 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structures of an engineered phospholipase D with specificity for secondary alcohol transphosphatidylation: insights into plasticity of substrate binding and activation.
Biochem.J., 478, 2021
|
|
7JRV
 
 | | Phospholipase D engineered mutant bound to phosphatidic acid (30 minute soak) | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dibutanoate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Vrielink, A, Samantha, A. | | Deposit date: | 2020-08-13 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structures of an engineered phospholipase D with specificity for secondary alcohol transphosphatidylation: insights into plasticity of substrate binding and activation.
Biochem.J., 478, 2021
|
|
7JS5
 
 | |
7JRW
 
 | | Phospholipase D engineered mutant bound to phosphatidic acid (5 day soak) | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dibutanoate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Vrielink, A, Samantha, A. | | Deposit date: | 2020-08-13 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structures of an engineered phospholipase D with specificity for secondary alcohol transphosphatidylation: insights into plasticity of substrate binding and activation.
Biochem.J., 478, 2021
|
|
2PZN
 
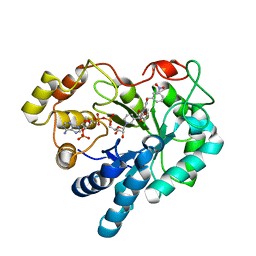 | | The crystallographic structure of Aldose Reductase IDD393 complex confirms Leu300 as a specificity determinant | | Descriptor: | (5-CHLORO-2-{[(3-NITROBENZYL)AMINO]CARBONYL}PHENOXY)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Ruiz, F, Hazemann, I, Darmanin, C, Mitschler, A, Van Zandt, M, Joachimiak, A, El-Kabbani, O, Podjarny, A. | | Deposit date: | 2007-05-18 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The Crystallographic Structure of Alr2-Idd393 Complex Confirms Leu300 as a Specificity Determinant
To be Published
|
|
7JS7
 
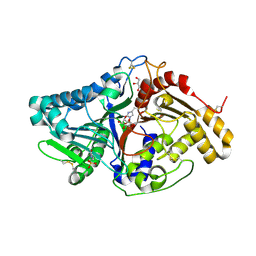 | |
7JRU
 
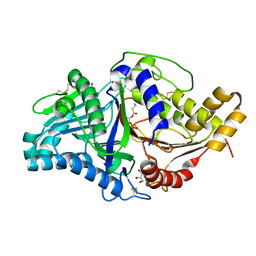 | | Phospholipase D engineered mutant bound to phosphatidic acid (8 hour soak) | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dibutanoate, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Vrielink, A, Samantha, A. | | Deposit date: | 2020-08-13 | | Release date: | 2021-04-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structures of an engineered phospholipase D with specificity for secondary alcohol transphosphatidylation: insights into plasticity of substrate binding and activation.
Biochem.J., 478, 2021
|
|
5VGC
 
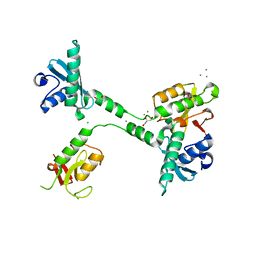 | | Crystal structure of the NleG5-1 effector (C200A) from Escherichia coli O157:H7 str. Sakai | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Borek, D, Valleau, D, Skarina, T, Jobin, M.C, Wawrzak, Z, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-04-10 | | Release date: | 2018-04-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the NleG5-1 effector (C200A) from Escherichia coli O157:H7 str. Sakai
To Be Published
|
|
4PIR
 
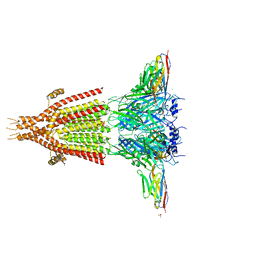 | | X-ray structure of the mouse serotonin 5-HT3 receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 5-hydroxytryptamine receptor 3A, ... | | Authors: | Hassaine, G, Deluz, C, Grasso, L, Wyss, R, Tol, M.B, Hovius, R, Graff, A, Stahlberg, H, Tomizaki, T, Desmyter, A, Moreau, C, Li, X.-D, Poitevin, F, Vogel, H, Nury, H. | | Deposit date: | 2014-05-09 | | Release date: | 2014-08-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | X-ray structure of the mouse serotonin 5-HT3 receptor.
Nature, 512, 2014
|
|
5A80
 
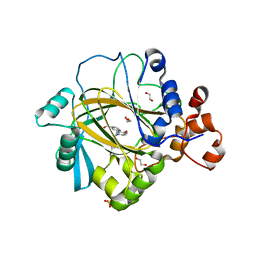 | | Crystal structure of human JMJD2A in complex with compound 40 | | Descriptor: | 1,2-ETHANEDIOL, 2-[5-[2-(3-methoxyphenyl)ethanoylamino]-2-oxidanyl-phenyl]pyridine-4-carboxylic acid, LYSINE-SPECIFIC DEMETHYLASE 4A, ... | | Authors: | Nowak, R, Velupillai, S, Krojer, T, Gileadi, C, Johansson, C, Korczynska, M, Le, D.D, Younger, N, Gregori-Puigjane, E, Tumber, A, Iwasa, E, Pollock, S.B, Ortiz Torres, I, Kopec, J, Tallant, C, Froese, S, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Shoichet, B.K, Fujimori, D.G, Oppermann, U. | | Deposit date: | 2015-07-11 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Docking and Linking of Fragments to Discover Jumonji Histone Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
2Q1Q
 
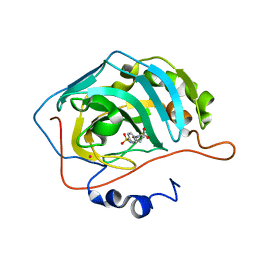 | | Carbonic anhydrase inhibitors. Interaction of the antiepileptic drug sulthiame with twelve mammalian isoforms: kinetic and X-Ray crystallographic studies | | Descriptor: | Carbonic anhydrase 2, MERCURY (II) ION, SULTHIAME, ... | | Authors: | Temperini, C, Innocenti, A, Mastrolorenzo, A, Scozzafava, A, Supuran, C.T. | | Deposit date: | 2007-05-25 | | Release date: | 2007-09-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Carbonic anhydrase inhibitors. Interaction of the antiepileptic drug sulthiame with twelve mammalian isoforms: Kinetic and X-ray crystallographic studies.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
1MZP
 
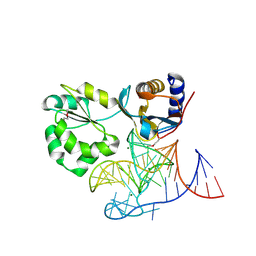 | | Structure of the L1 protuberance in the ribosome | | Descriptor: | 50s ribosomal protein L1P, MAGNESIUM ION, fragment of 23S rRNA | | Authors: | Nikulin, A, Eliseikina, I, Tishchenko, S, Nevskaya, N, Davydova, N, Platonova, O, Piendl, W, Selmer, M, Liljas, A, Zimmermann, R, Garber, M, Nikonov, S. | | Deposit date: | 2002-10-09 | | Release date: | 2003-01-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the L1 protuberance in the ribosome.
Nat.Struct.Biol., 10, 2003
|
|
1MZ4
 
 | | Crystal Structure of Cytochrome c550 from Thermosynechococcus elongatus | | Descriptor: | BICARBONATE ION, GLYCEROL, HEME C, ... | | Authors: | Kerfeld, C.A, Sawaya, M.R, Bottin, H, Tran, K.T, Sugiura, M, Kirilovsky, D, Krogmann, D, Yeates, T.O, Boussac, A. | | Deposit date: | 2002-10-05 | | Release date: | 2003-09-23 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and EPR characterization of the soluble form of cytochrome c-550 and of the psbV2 gene product from the cyanobacterium Thermosynechococcus elongatus.
Plant Cell.Physiol., 44, 2003
|
|
4JFL
 
 | | Increasing the Efficiency Efficiency of Ligands for the FK506-Binding Protein 51 by Conformational Control: Complex of FKBP51 with 6-({(1S,5R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-2-oxo-3,9-diazabicyclo[3.3.1]non-9-yl}sulfonyl)-1,3-benzothiazol-2(3H)-one | | Descriptor: | 6-({(1S,5R)-3-[2-(3,4-dimethoxyphenoxy)ethyl]-2-oxo-3,9-diazabicyclo[3.3.1]non-9-yl}sulfonyl)-1,3-benzothiazol-2(3H)-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Wang, Y, Kirschner, A, Fabian, A, Gopalakrishnan, R, Kress, C, Hoogeland, B, Koch, U, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2013-02-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Increasing the efficiency of ligands for FK506-binding protein 51 by conformational control.
J.Med.Chem., 56, 2013
|
|
4APN
 
 | | Structure of TR from Leishmania infantum in complex with a diarylpirrole-based inhibitor | | Descriptor: | 4-[[1-(4-ethylphenyl)-2-methyl-5-(4-methylsulfanylphenyl)pyrrol-3-yl]methyl]thiomorpholine, FLAVIN-ADENINE DINUCLEOTIDE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Baiocco, P, Ilari, A, Colotti, G, Biava, M. | | Deposit date: | 2012-04-04 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Inhibition of Leishmania Infantum Trypanothione Reductase by Azole-Based Compounds: A Comparative Analysis with its Physiological Substrate by X-Ray Crystallography.
Chemmedchem, 8, 2013
|
|
2JIS
 
 | | Human cysteine sulfinic acid decarboxylase (CSAD) in complex with PLP. | | Descriptor: | CYSTEINE SULFINIC ACID DECARBOXYLASE, NITRATE ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Collins, R, Moche, M, Arrowsmith, C, Berglund, H, Busam, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Welin, M, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-30 | | Release date: | 2007-08-28 | | Last modified: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of Human Cysteine Sulfinic Acid Decarboxylase (Csad)
To be Published
|
|
4PU2
 
 | | Crystal structure of Aminopeptidase N in complex with the phosphonic acid analogue of leucine L-(R)-LeuP | | Descriptor: | Aminopeptidase N, GLYCEROL, LEUCINE PHOSPHONIC ACID, ... | | Authors: | Nocek, B, Vassiliou, S, Berlicki, L, Mulligan, R, Mucha, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2014-03-11 | | Release date: | 2014-06-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.095 Å) | | Cite: | Crystal structure of Aminopeptidase N in complex with the phosphonic acid analogue of leucine
To be Published
|
|
5VF6
 
 | |
4PED
 
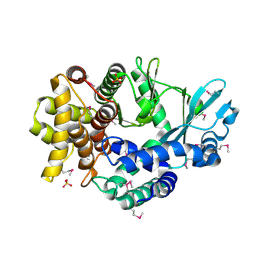 | | Mitochondrial ADCK3 employs an atypical protein kinase-like fold to enable coenzyme Q biosynthes | | Descriptor: | Chaperone activity of bc1 complex-like, mitochondrial, SULFATE ION | | Authors: | Bingman, C.A, Smith, R, Joshi, S, Stefely, J.A, Reidenbach, A.G, Ulbrich, A, Oruganty, O, Floyd, B.J, Jochem, A, Saunders, J.M, Johnson, I.E, Wrobel, R.L, Barber, G.E, Lee, D, Li, S, Kannan, N, Coon, J.J, Pagliarini, D.J, Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2014-04-22 | | Release date: | 2014-11-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Mitochondrial ADCK3 Employs an Atypical Protein Kinase-like Fold to Enable Coenzyme Q Biosynthesis.
Mol.Cell, 57, 2015
|
|
1N3T
 
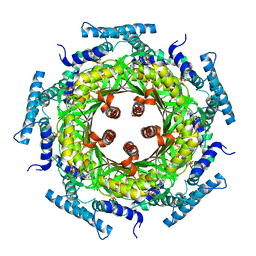 | | Biosynthesis of pteridins. Reaction mechanism of GTP cyclohydrolase I | | Descriptor: | GTP cyclohydrolase I, GUANOSINE-5'-TRIPHOSPHATE | | Authors: | Rebelo, J, Auerbach, G, Bader, G, Bracher, A, Nar, H, Hoesl, C, Schramek, N, Kaiser, J, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-10-29 | | Release date: | 2003-10-14 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Biosynthesis of Pteridines. Reaction Mechanism of GTP Cyclohydrolase I
J.MOL.BIOL., 326, 2003
|
|
1MQS
 
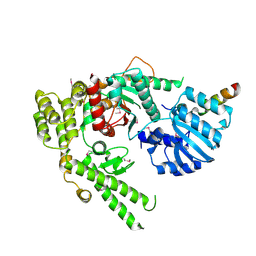 | |
2QIB
 
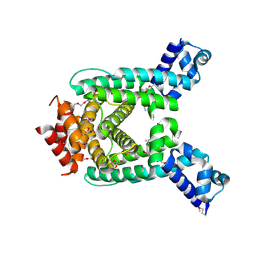 | | Crystal structure of tetR-family transcriptional regulator from Streptomyces coelicolor | | Descriptor: | HEXAETHYLENE GLYCOL, tetR-family transcriptional regulator | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-07-03 | | Release date: | 2007-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The crystal structure of tetR-family transcriptional regulator from Streptomyces coelicolor.
To be Published
|
|
1MSZ
 
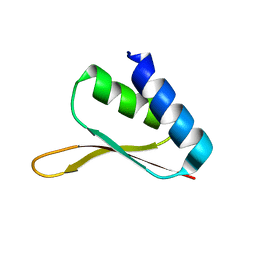 | | Solution structure of the R3H domain from human Smubp-2 | | Descriptor: | DNA-binding protein SMUBP-2 | | Authors: | Liepinsh, E, Leonchiks, A, Sharipo, A, Guignard, L, Otting, G. | | Deposit date: | 2002-09-20 | | Release date: | 2002-10-09 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the R3H domain from human Smubp-2
J.Mol.Biol., 326, 2003
|
|
2YPJ
 
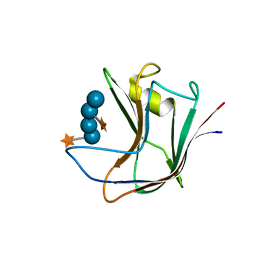 | | Non-catalytic carbohydrate binding module CBM65B | | Descriptor: | ENDOGLUCANASE CEL5A, alpha-D-xylopyranose-(1-6)-beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-[alpha-D-xylopyranose-(1-6)]beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Luis, A.S, Venditto, I, Prates, J.A.M, Ferreira, L.M.A, Temple, M.J, Rogowski, A, Basle, A, Xue, J, Knox, J.P, Gilbert, H.J, Fontes, C.M.G.A, Najmudin, S. | | Deposit date: | 2012-10-30 | | Release date: | 2012-12-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Understanding How Non-Catalytic Carbohydrate Binding Modules Can Display Specificity for Xyloglucan
J.Biol.Chem., 288, 2013
|
|
1MWW
 
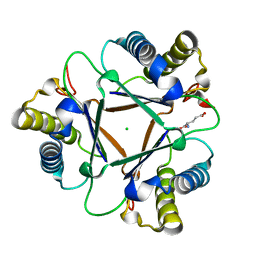 | | THE STRUCTURE OF THE HYPOTHETICAL PROTEIN HI1388.1 FROM HAEMOPHILUS INFLUENZAE REVEALS A TAUTOMERASE/MIF FOLD | | Descriptor: | CHLORIDE ION, GLUTAMIC ACID, HYPOTHETICAL PROTEIN HI1388.1 | | Authors: | Lehmann, C, Pullalarevu, S, Krajewski, W, Galkin, A, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2002-10-01 | | Release date: | 2003-11-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structure of the Hypothetical Protein HI1388.1 from Haemophilus influenzae
To be Published
|
|
